MBI Videos
Examining the dynamics across network space using Database for Dynamics
-
 Tomas Gedeon
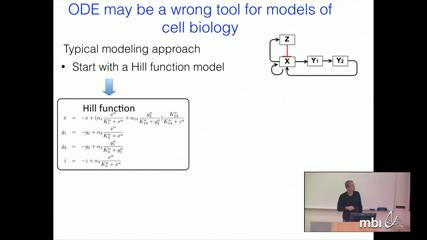
Tomas GedeonExperimental data on gene regulation is mostly qualitative, where the only information available about pairwise interactions is the presence of either up-or down- regulation. Quantitative data is often subject to large uncertainty and is mostly in terms of fold differences. Given these realities, it is very diffcult to make reliable predictions using mathematical models. The current approach of choosing reasonable parameter values, a few initial conditions and then making predictions based on resulting solutions is severely subsampling both the parameter and phase space. This approach does not produce provable and reliable predictions. We present a new approach that uses continuous time Boolean networks as a platform for qualitative studies of gene regulation. We compute a Database for Dynamics, which rigorously approximates global dynamics over entire parameter space. The results obtained by this method provably capture the dynamics at a predetermined spatial scale. We apply our approach to study neighborhood of a given network in the space of networks. We start with a E2F-Rb network underlying the mammalian cell cycle restriction point and show that majority of the parameters support either the GO, NO-GO, or bistability between these two states. We then sample perturbations of this network and study robustness of this dynamics in the network space.
