-

Graham Coop
Parallel mutations and partial sweeps
-

Thomas Mailund
A coalescent process Markov in time and space
-

Carolin Kosiol
In the past, two kinds of Markov models have been considered to describe protein sequence evolution. Codon-level models have been mechanistic, with a small number of parameters designed to take into account features such as transition-transversion bias, codon frequency bias and synonymous-nonsynonymous amino acid substitution bias. Amino acid models have been empirical, attempting to summarize the replacement patterns observed in large quantities of data and not explicitly considering the distinct factors that shape protein evolution.
We have estimated the first empirical codon model using Maximum Likelihood techniques. Our results show that modelling the evolutionary process is improved by allowing for single, double and triple nucleotide changes; the affiliation between DNA triplets and the amino acid they encode is a main factor driving evolution; and the nonsynonymous-synonymous rate ratio is a suitable measure to classify substitution patterns observed for different proteins. However, are the double and triple changes instantaneous? We aim to take advantage of newly available re-sequencing data to further improve our models and understanding of the evolution of protein coding sequences. Specifically, we estimate empirical codon substitution models from re-sequencing data of several Drosophila species.
-

Ana Arribas-Gil
Models for pairwise alignment based on the TKF (Thorne, Kishino and Felsenstein 1991) indel process fit into the pair-Hidden Markov Model (pair-HMM). Observations in a pair-HMM are formed by the couple of sequences to be aligned and the hidden alignment is a Markov chain. Many efficient algorithms have been developed to estimate alignments and evolution parameters in this context. From a theoretical point of view, it is also interesting to investigate the statistical properties of the estimators computed by these algorithms. In particular, we will discuss consistency of maximum likelihood and Bayesian estimators. In the context of multiple alignment, the classical indel evolution process for the sequences (TKF) provides a complex hidden variable model for the alignment in which the phylogenetic relationships between the sequences must be taken into account. We provide a theoretical framework for this model and study, as for the pairwise alignment, the consistency of estimators.
-
Marc Suchard
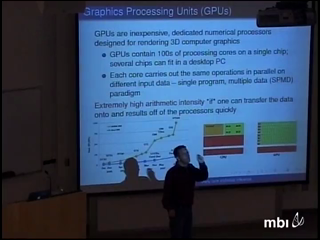
Massive numerical integration plagues the statistical inference of partially observed stochastic processes. An important biological example entertains partially observed continuous-time Markov chains (CTMCs) to model molecular sequence evolution. Joint inference of phylogenetic trees and codon-based substitution models of sequence evolution remains computationally impractical. Parallelizing data likelihood calculations is an obvious strategy; however, across a cluster-computer, this scales with the total number of processing cores, incurring considerable cost to achieve reasonable run-time.
To solve this problem, I describe many-core computing algorithms that harness inexpensive graphics processing units (GPUs) for calculation of the likelihood under CTMC models of evolution. High-end GPUs containing hundreds of cores and are low-cost. These novel algorithms are particularly efficient for large state-spaces, including codon models, and large data sets, such as full genome alignments where we demonstrate up to 150-fold speed-up. I conclude with a discussion of the future of many-core computing in statistics and touch upon recent experiences with massively large and high-dimensional mixture models.
-

Cedric Feschotte
I will provide an overview of our studies of the evolutionary dynamics of transposable elements and their impact on eukaryotic evolution. The emphasis will be on class 2 or DNA transposons in mammalian genomes. Genomic analyses reveal that a vast diversity of DNA transposons exists in mammalian genomes and that large cohorts of elements have been integrated in a lineage-specific fashion, generating extensive genomic variation among and within the major lineages of placental mammals. The evolutionary dynamics of DNA transposons in mammals is characterized by repeated episodes of horizontal transfer across widely diverged species, a mode of transmission that appears to be facilitated by blood-borne invertebrate parasites. Through several examples, I will illustrate how DNA transposons have been a recurrent source of genetic innovation throughout mammalian evolution, contributing to the shuffling of pre-existing genes and to the emergence of new genes and concomitantly to the assembly of lineage-specific regulatory circuits.
-
Daniel Falush
Inference of population structure using dense genotype data
-
Anna Kedzierska
Phylogenetic data arising on different tree topologies might be mixed on a given alignment. Models taking into it account usually contain a large number of parameters and the usual tests for model fitting cannot deal with them. Here we discuss an approach for model fitting of algebraic models on m-tree mixtures. This is joint work in progress with Marta Casanellas and Jesus Fernandez-Sanchez
-

Adam Siepel
Complete genome sequences are now available for individuals representing several distinct human populations. The primary motivation for collecting these sequences has been to characterize human diversity, facilitate disease association studies, and pave the way for an era of "personalized medicine". However, these data also contain valuable information about human evolution. Here I will describe an effort currently in progress to estimate evolutionary parameters such as the times at which major population groups diverged and the effective sizes of ancestral human populations, based on sequence data for two West Africans, two East Asians, two individuals of European descent, and a Khoisan hunter gatherer from the Kalahari Desert in Southern Africa. This work involves an interesting mixture of traditional phylogenetics and population genetics, and also requires a number of challenging technical issues -- concerning alignment and sequencing error, and genotype estimation -- to be addressed. I will describe our statistical model, which is derived from the MCMCCOAL model by Rannala and Yang, and our Markov chain Monte Carlo methods for inference. I will also present preliminary results from our analysis and discuss their relationship with what is currently known about early human evolution.
-
Matthew Rasmussen
Advances in genome sequencing have enabled the study of evolution across both genomes and large clades of species, and has been especially useful for studying gene families as they expand and contract over evolutionary time by gene duplication and loss events. Here, we present a new approach for the reconstruction of gene-tree phylogenies that models simultaneously gene and species evolution, which enables significantly increased reconstruction accuracy by incorporating genome-wide information. We introduce SPIDIR v2, a Bayesian method for gene tree reconstruction that incorporates within a unified framework models for gene duplication and loss, gene- and species-specific rate variation, and sequence substitution. The method includes an Expectation-Maximization method for inferring the distribution of species-specific and gene-specific rates across unambiguous orthologs, which we then use as priors in the reconstruction of any family. We have also developed a novel fast search algorithm that uses the Birth-Death process to more efficiently explore tree space leading to faster and more accurate recovery of a Maximum a posteriori (MAP) gene tree. We have applied our method in two clades of fully-sequenced genomes consisting of 12 Drosophila and 16 fungal species as well as simulated data. We use these extensive benchmarks to study the overall and branch-level accuracy for SPIDIR and many other phylogenetic methods. We find that SPIDIR achieves dramatic improvements in reconstruction accuracy over both traditional phylogenetic methods (Maximum Parsimony, Maximum Likelihood, and Bayesian methods) as well as other species-aware methods (SYNERGY, PRIME-GSR) in both real and simulated datasets.
Work done in collaboration with Manolis Kelli.
-

David Liberles
In Darwinian evolution, mutations occur approximately at random in a gene, turned into amino acid mutations by the genetic code. Some mutations are fixed to become substitutions and some are eliminated from the population. Partitioning pairs of closely related species with complete genome sequences by population size, we look at the PAM matrices generated for these partitions and compare the substitution patterns between species. A population genetic model is generated that relates the relative fixation probabilities of different types of mutations to the selective pressure and population size. Parameterizations of the average and distribution of selective pressures for different amino acid substitution types in different population size comparisons are generated using a Bayesian framework. We find that partitions in population size as well as in substitution type are required to explain the substitution data. Mechanistic explanations of this will be discussed.
In a second line of work, models for gene duplication will be discussed, with potential extension to the Gene tree/species tree reconciliation problem. This second trajectory involves an extension of birth death models based upon an exponential distribution using a generalization based upon a Weibull distribution. Distinct parameterizations of the Weibull distribution are expected to be consistent with the neofunctionalization, subfunctionalization, and dosage compensation processes. Ongoing work involves the extension of the Weibull-based models to the gene tree/species tree reconciliation approach using a Bayesian framework, with potential utility in additionally identifying underlying evolutionary processes.
 Graham CoopParallel mutations and partial sweeps
Graham CoopParallel mutations and partial sweeps Thomas MailundA coalescent process Markov in time and space
Thomas MailundA coalescent process Markov in time and space Carolin KosiolIn the past, two kinds of Markov models have been considered to describe protein sequence evolution. Codon-level models have been mechanistic, with a small number of parameters designed to take into account features such as transition-transversion bias, codon frequency bias and synonymous-nonsynonymous amino acid substitution bias. Amino acid models have been empirical, attempting to summarize the replacement patterns observed in large quantities of data and not explicitly considering the distinct factors that shape protein evolution.
Carolin KosiolIn the past, two kinds of Markov models have been considered to describe protein sequence evolution. Codon-level models have been mechanistic, with a small number of parameters designed to take into account features such as transition-transversion bias, codon frequency bias and synonymous-nonsynonymous amino acid substitution bias. Amino acid models have been empirical, attempting to summarize the replacement patterns observed in large quantities of data and not explicitly considering the distinct factors that shape protein evolution. Ana Arribas-GilModels for pairwise alignment based on the TKF (Thorne, Kishino and Felsenstein 1991) indel process fit into the pair-Hidden Markov Model (pair-HMM). Observations in a pair-HMM are formed by the couple of sequences to be aligned and the hidden alignment is a Markov chain. Many efficient algorithms have been developed to estimate alignments and evolution parameters in this context. From a theoretical point of view, it is also interesting to investigate the statistical properties of the estimators computed by these algorithms. In particular, we will discuss consistency of maximum likelihood and Bayesian estimators. In the context of multiple alignment, the classical indel evolution process for the sequences (TKF) provides a complex hidden variable model for the alignment in which the phylogenetic relationships between the sequences must be taken into account. We provide a theoretical framework for this model and study, as for the pairwise alignment, the consistency of estimators.
Ana Arribas-GilModels for pairwise alignment based on the TKF (Thorne, Kishino and Felsenstein 1991) indel process fit into the pair-Hidden Markov Model (pair-HMM). Observations in a pair-HMM are formed by the couple of sequences to be aligned and the hidden alignment is a Markov chain. Many efficient algorithms have been developed to estimate alignments and evolution parameters in this context. From a theoretical point of view, it is also interesting to investigate the statistical properties of the estimators computed by these algorithms. In particular, we will discuss consistency of maximum likelihood and Bayesian estimators. In the context of multiple alignment, the classical indel evolution process for the sequences (TKF) provides a complex hidden variable model for the alignment in which the phylogenetic relationships between the sequences must be taken into account. We provide a theoretical framework for this model and study, as for the pairwise alignment, the consistency of estimators. Marc SuchardMassive numerical integration plagues the statistical inference of partially observed stochastic processes. An important biological example entertains partially observed continuous-time Markov chains (CTMCs) to model molecular sequence evolution. Joint inference of phylogenetic trees and codon-based substitution models of sequence evolution remains computationally impractical. Parallelizing data likelihood calculations is an obvious strategy; however, across a cluster-computer, this scales with the total number of processing cores, incurring considerable cost to achieve reasonable run-time.
Marc SuchardMassive numerical integration plagues the statistical inference of partially observed stochastic processes. An important biological example entertains partially observed continuous-time Markov chains (CTMCs) to model molecular sequence evolution. Joint inference of phylogenetic trees and codon-based substitution models of sequence evolution remains computationally impractical. Parallelizing data likelihood calculations is an obvious strategy; however, across a cluster-computer, this scales with the total number of processing cores, incurring considerable cost to achieve reasonable run-time. Cedric FeschotteI will provide an overview of our studies of the evolutionary dynamics of transposable elements and their impact on eukaryotic evolution. The emphasis will be on class 2 or DNA transposons in mammalian genomes. Genomic analyses reveal that a vast diversity of DNA transposons exists in mammalian genomes and that large cohorts of elements have been integrated in a lineage-specific fashion, generating extensive genomic variation among and within the major lineages of placental mammals. The evolutionary dynamics of DNA transposons in mammals is characterized by repeated episodes of horizontal transfer across widely diverged species, a mode of transmission that appears to be facilitated by blood-borne invertebrate parasites. Through several examples, I will illustrate how DNA transposons have been a recurrent source of genetic innovation throughout mammalian evolution, contributing to the shuffling of pre-existing genes and to the emergence of new genes and concomitantly to the assembly of lineage-specific regulatory circuits.
Cedric FeschotteI will provide an overview of our studies of the evolutionary dynamics of transposable elements and their impact on eukaryotic evolution. The emphasis will be on class 2 or DNA transposons in mammalian genomes. Genomic analyses reveal that a vast diversity of DNA transposons exists in mammalian genomes and that large cohorts of elements have been integrated in a lineage-specific fashion, generating extensive genomic variation among and within the major lineages of placental mammals. The evolutionary dynamics of DNA transposons in mammals is characterized by repeated episodes of horizontal transfer across widely diverged species, a mode of transmission that appears to be facilitated by blood-borne invertebrate parasites. Through several examples, I will illustrate how DNA transposons have been a recurrent source of genetic innovation throughout mammalian evolution, contributing to the shuffling of pre-existing genes and to the emergence of new genes and concomitantly to the assembly of lineage-specific regulatory circuits. Daniel FalushInference of population structure using dense genotype data
Daniel FalushInference of population structure using dense genotype data Anna KedzierskaPhylogenetic data arising on different tree topologies might be mixed on a given alignment. Models taking into it account usually contain a large number of parameters and the usual tests for model fitting cannot deal with them. Here we discuss an approach for model fitting of algebraic models on m-tree mixtures. This is joint work in progress with Marta Casanellas and Jesus Fernandez-Sanchez
Anna KedzierskaPhylogenetic data arising on different tree topologies might be mixed on a given alignment. Models taking into it account usually contain a large number of parameters and the usual tests for model fitting cannot deal with them. Here we discuss an approach for model fitting of algebraic models on m-tree mixtures. This is joint work in progress with Marta Casanellas and Jesus Fernandez-Sanchez Adam SiepelComplete genome sequences are now available for individuals representing several distinct human populations. The primary motivation for collecting these sequences has been to characterize human diversity, facilitate disease association studies, and pave the way for an era of "personalized medicine". However, these data also contain valuable information about human evolution. Here I will describe an effort currently in progress to estimate evolutionary parameters such as the times at which major population groups diverged and the effective sizes of ancestral human populations, based on sequence data for two West Africans, two East Asians, two individuals of European descent, and a Khoisan hunter gatherer from the Kalahari Desert in Southern Africa. This work involves an interesting mixture of traditional phylogenetics and population genetics, and also requires a number of challenging technical issues -- concerning alignment and sequencing error, and genotype estimation -- to be addressed. I will describe our statistical model, which is derived from the MCMCCOAL model by Rannala and Yang, and our Markov chain Monte Carlo methods for inference. I will also present preliminary results from our analysis and discuss their relationship with what is currently known about early human evolution.
Adam SiepelComplete genome sequences are now available for individuals representing several distinct human populations. The primary motivation for collecting these sequences has been to characterize human diversity, facilitate disease association studies, and pave the way for an era of "personalized medicine". However, these data also contain valuable information about human evolution. Here I will describe an effort currently in progress to estimate evolutionary parameters such as the times at which major population groups diverged and the effective sizes of ancestral human populations, based on sequence data for two West Africans, two East Asians, two individuals of European descent, and a Khoisan hunter gatherer from the Kalahari Desert in Southern Africa. This work involves an interesting mixture of traditional phylogenetics and population genetics, and also requires a number of challenging technical issues -- concerning alignment and sequencing error, and genotype estimation -- to be addressed. I will describe our statistical model, which is derived from the MCMCCOAL model by Rannala and Yang, and our Markov chain Monte Carlo methods for inference. I will also present preliminary results from our analysis and discuss their relationship with what is currently known about early human evolution. Matthew RasmussenAdvances in genome sequencing have enabled the study of evolution across both genomes and large clades of species, and has been especially useful for studying gene families as they expand and contract over evolutionary time by gene duplication and loss events. Here, we present a new approach for the reconstruction of gene-tree phylogenies that models simultaneously gene and species evolution, which enables significantly increased reconstruction accuracy by incorporating genome-wide information. We introduce SPIDIR v2, a Bayesian method for gene tree reconstruction that incorporates within a unified framework models for gene duplication and loss, gene- and species-specific rate variation, and sequence substitution. The method includes an Expectation-Maximization method for inferring the distribution of species-specific and gene-specific rates across unambiguous orthologs, which we then use as priors in the reconstruction of any family. We have also developed a novel fast search algorithm that uses the Birth-Death process to more efficiently explore tree space leading to faster and more accurate recovery of a Maximum a posteriori (MAP) gene tree. We have applied our method in two clades of fully-sequenced genomes consisting of 12 Drosophila and 16 fungal species as well as simulated data. We use these extensive benchmarks to study the overall and branch-level accuracy for SPIDIR and many other phylogenetic methods. We find that SPIDIR achieves dramatic improvements in reconstruction accuracy over both traditional phylogenetic methods (Maximum Parsimony, Maximum Likelihood, and Bayesian methods) as well as other species-aware methods (SYNERGY, PRIME-GSR) in both real and simulated datasets.
Matthew RasmussenAdvances in genome sequencing have enabled the study of evolution across both genomes and large clades of species, and has been especially useful for studying gene families as they expand and contract over evolutionary time by gene duplication and loss events. Here, we present a new approach for the reconstruction of gene-tree phylogenies that models simultaneously gene and species evolution, which enables significantly increased reconstruction accuracy by incorporating genome-wide information. We introduce SPIDIR v2, a Bayesian method for gene tree reconstruction that incorporates within a unified framework models for gene duplication and loss, gene- and species-specific rate variation, and sequence substitution. The method includes an Expectation-Maximization method for inferring the distribution of species-specific and gene-specific rates across unambiguous orthologs, which we then use as priors in the reconstruction of any family. We have also developed a novel fast search algorithm that uses the Birth-Death process to more efficiently explore tree space leading to faster and more accurate recovery of a Maximum a posteriori (MAP) gene tree. We have applied our method in two clades of fully-sequenced genomes consisting of 12 Drosophila and 16 fungal species as well as simulated data. We use these extensive benchmarks to study the overall and branch-level accuracy for SPIDIR and many other phylogenetic methods. We find that SPIDIR achieves dramatic improvements in reconstruction accuracy over both traditional phylogenetic methods (Maximum Parsimony, Maximum Likelihood, and Bayesian methods) as well as other species-aware methods (SYNERGY, PRIME-GSR) in both real and simulated datasets. David LiberlesIn Darwinian evolution, mutations occur approximately at random in a gene, turned into amino acid mutations by the genetic code. Some mutations are fixed to become substitutions and some are eliminated from the population. Partitioning pairs of closely related species with complete genome sequences by population size, we look at the PAM matrices generated for these partitions and compare the substitution patterns between species. A population genetic model is generated that relates the relative fixation probabilities of different types of mutations to the selective pressure and population size. Parameterizations of the average and distribution of selective pressures for different amino acid substitution types in different population size comparisons are generated using a Bayesian framework. We find that partitions in population size as well as in substitution type are required to explain the substitution data. Mechanistic explanations of this will be discussed.
David LiberlesIn Darwinian evolution, mutations occur approximately at random in a gene, turned into amino acid mutations by the genetic code. Some mutations are fixed to become substitutions and some are eliminated from the population. Partitioning pairs of closely related species with complete genome sequences by population size, we look at the PAM matrices generated for these partitions and compare the substitution patterns between species. A population genetic model is generated that relates the relative fixation probabilities of different types of mutations to the selective pressure and population size. Parameterizations of the average and distribution of selective pressures for different amino acid substitution types in different population size comparisons are generated using a Bayesian framework. We find that partitions in population size as well as in substitution type are required to explain the substitution data. Mechanistic explanations of this will be discussed.