MBI Videos
Workshop 1: Family-based Genomic Studies
-
 Pamela Brock
Pamela BrockAbstract not available
-
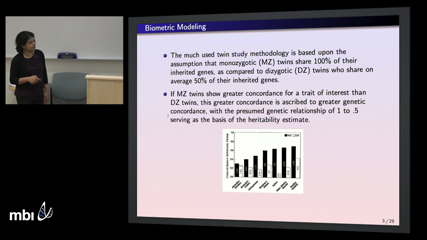 Saonli Basu
Saonli BasuThe development of a complex disease is an intricate interplay of genetic and environmental factors. "Heritability" is defined as the proportion of total trait variance due to genetic factors within a given population. Studies with monozygotic and dizygotic twins allow us to estimate heritability by fitting an "ACE" model which estimates the proportion of trait variance explained by additive genetic (A), common shared environment (C), and unique non-shared environmental (E) latent effects, thus helping us better understand disease risk and etiology. IIn this paper, we develop a flexible generalized estimating equations framework (``GEE2'') for fitting twin ACE models that requires minimal distributional assumptions; rather only the first two moments need to be correctly specified. We show that two commonly used methods for estimating heritability, the normal ACE model (``NACE'') and Falconer's method, can both be fit within this unified GEE2 framework, which additionally provides robust standard errors. Although the traditional Falconer's method cannot directly adjust for covariates, the corresponding GEE2 version (``GEE2-Falconer'') can incorporate covarimate effects (e.g. let heritability vary by sex or age). Given non-normal data, these GEE2 models attain significantly better coverage of the true heritability compared to the traditional NACE and Falconer's methods. Finally, we demonstrate that Falconer's method can consistently estimate heritability when the ACE variance parameters differ between MZ and DZ twins; whereas the NACE will produce biased estimates in such settings.
Joint work with Jaron Arbet, Department of Biostatistics, University of Minnesota
-
 Meng Wang
Meng WangMotivation: In recent years, there has been an increasing interest in using common single-nucleotide polymorphisms (SNPs) amassed in genome-wide association studies to investigate rare haplotype effects on complex diseases. Evidence has suggested that rare haplotypes may tag rare causal single-nucleotide variants, making SNP-based rare haplotype analysis not only cost effective, but also more valuable for detecting causal variants. Although a number of methods for detecting rare haplotype association have been proposed in recent years, they are population based and thus susceptible to population stratification.
Results: We propose family-triad-based logistic Bayesian Lasso (famLBL) for estimating effects of haplotypes on complex diseases using SNP data. By choosing appropriate prior distribution, effect sizes of unassociated haplotypes can be shrunk toward zero, allowing for more precise estimation of associated haplotypes, especially those that are rare, thereby achieving greater detection power. We evaluate famLBL using simulation to gauge its type I error and power. Compared with its population counterpart, LBL, highlights famLBL’s robustness property in the presence of population substructure. Further investigation by comparing famLBL with Family-Based Association Test (FBAT) reveals its advantage for detecting rare haplotype association.
-
 Alyssa Clay-Gilmour
Alyssa Clay-GilmourMultiple myeloma (MM) is a result of a malignant transformation of plasma cells that is preceded by the presence of an asymptomatic clonal plasma cell expansion, a condition referred to as monoclonal gammopathy of undetermined significance (MGUS). We and others have shown familial aggregation of MM and MGUS. Evidence from epidemiologic, family and genome-wide association studies (GWAS) suggests a genetic component underlying MM etiology. GWAS have successfully established 17 common genetic risk loci for MM to date and recently, rare inherited susceptibility variants in the LSD1 / KDM1A and USP45 genes were identified in familial MM / MGUS kindreds. Family-based approaches may be used to elucidate genetic variation contributing to familial MM. Genetic linkage analysis has historically been used to detect the chromosomal location of disease genes. The objective of this study was to conduct a linkage analysis of MM / MGUS families to identify genomic regions for MM / MGUS.
-
 Jonathan Haines
Jonathan HainesGenetic studies of diseases of aging have been done predominantly in clinic-based case-control datasets drawn from the general population. While these have the advantage of being relatively easy to collect and thus can generate large sample sizes, they do have limitations in ascertainment bias, differences in case and control ascertainment, and focus on genetic association analyses. Using special populations, such as the mid-Western Amish, overcomes several of these limitations.
Over the past 15 years, we have worked collaboratively to collect phenotype and genotype information on the Amish of Holmes county in Ohio, and Elkhart, LaGrange, and Adams counties in Indiana. The Amish are culturally and genetically isolated and their lifestyle tends to be quite homogeneous, making genetic studies quite valuable. We have focused our efforts on two significant diseases of aging: Alzheimer disease (AD) and Age Related Macular Degeneration (AMD). Our ongoing studies have demonstrated that the genetic architecture of Ad and AMD differ significantly from the general population, strongly suggesting that novel loci exist in the Amish. Current studies are aimed at finding these novel loci using a combination of genome wide association and whole genome sequencing data.
-
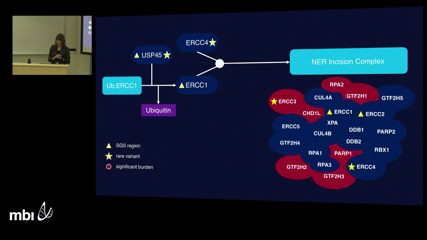 Rosalie Waller
Rosalie WallerHigh-risk pedigrees (HRPs) are a key design in mapping rare and highly-penetrant genes in Mendelian-like diseases. However, success with the HRP design in complex diseases has been modest, in part because standard methods do not adequately address genetic heterogeneity. Novel methods are needed to re-invigorate HRP designs for gene-discovery in complex diseases. Extended high-risk pedigrees can contain sufficient meioses to gain power for gene mapping as single pedigrees; however, intrafamilial heterogeneity may still exist. To address intrafamilial heterogeneity, we expanded on the Shared Genomic Segment (SGS) method, a large pedigree mapping method that identifies subsets of cases within an extended pedigree that share segregating chromosomal regions. Here, I will describe this strategy and our application to high-risk myeloma pedigrees.
-
 Daniel Koboldt
Daniel KoboldtChildren with rare inherited conditions are increasingly referred for clinical exome sequencing, which yields a positive finding in only ~25-35% of them. For the remaining as-yet-undiagnosed cases, research sequencing of the proband and available family members has the potential to uncover new genetic etiologies of disease. Our institute has enrolled more than 40 families suffering rare inherited conditions into a research genomics protocol. Using predominantly whole genome sequencing (WGS) of multiple family members, we have identified likely causal variants in 30% of cases and strong candidate variants in another 20%. Here, I describe the workflow of our rare disease genomics research program, including recruitment and case selection, sequencing/analysis strategies, candidate validation, and reporting of results. I will also highlight some solved cases whose underlying etiology or phenotypic association challenges the current knowledge of genotype-phenotype relationships.
-
 Christopher Bartlett
Christopher BartlettApplying quantum physics to build quantum devices for computing has recently become reality with companies such as Google, IBM, and Intel making prototypes for algorithm experimentation. These devices demonstrate that binary computing states (0 vs. 1) can be manipulated using the rules of quantum mechanics to include superposition, entanglement, and wave interference as fundamentally new avenues for computing algorithms. While quantum algorithms have already shown in-principle speed-ups over classical computation for certain classes of problems such as factoring prime numbers, finding new algorithms for statistical computation such as machine learning is ongoing. The key differences between classical and quantum computing will be discussed in the context addressing genomics questions through simple quantum machine learning examples.
-
 Catherine Stein
Catherine SteinTuberculosis (TB) remains a major public health threat globally, and several studies have demonstrated a role for human genetic factors underlying TB risk. However, exposure to the causal bacterium, Mycobacterium tuberculosis, is a necessary risk factor for TB, and few population-based studies appropriately account for this exposure. In this talk, I will describe how we’ve utilized a family study to examine the genetic epidemiology of TB and address limitations in the extant literature. I will present both key findings and future directions.
