MBI Videos
Workshop 3: Modeling and Analysis of Dynamic Social Networks
-
 Scott Duxbury
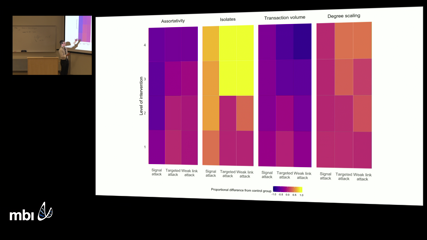
Scott DuxburyInterconnected systems in physical, biological, and social sciences are often at risk of attacks from exogenous sources. As a result, a growing number of studies focus on network attack tolerance. In general, this body of research assumes that damage to cross-sectional networks persists over time and has almost ubiquitously focused on attack strategies targeting integral vertices or edges. At issue, however, is that many networks are dynamic, especially in the social sciences, and capable of adaptive responses to attacks. Relatedly, network attack strategies may be diffuse, targeting an array of weak links, rather than high profile vertices. Together, these two omissions limit researchers’ ability to reach firm conclusions or derive policy recommendations from past research. Expanding on this prior work, we examine data collected from an online drug trafficking network comprised of roughly 7,400 actors and 17,000 illicit drug transactions observed over 14 months. We use these data to develop an agent-based simulation experiment evaluating how the drug trafficking network responds to targeted and diffuse attacks. Results show that the network suffers substantial damage from diffuse attacks and that conventional methods for evaluating network robustness do a poor job of representing this type of damage. In particular, cross-sectional measures of network robustness in the simulated output networks suggest that the networks actually grow stronger in the aftermath of a diffuse attack, despite losing a substantial portion of edges and vertices. Pertinent to policy, these results indicate that the diffuse attack strategy evaluated in this study is an effective tactic for curbing online drug trafficking—an issue which has vexed law enforcement for some time.
-
 John Light
John LightMany systems of scientific interest can be naturally thought of as networks of evolving relationships, for instance, linkages between neurons in the brain, computers on the internet, or adolescents in a high school. The last 20 years or so have seen rapid progress in methods for the empirical study of network evolution. The Stochastic Actor-Oriented Model (SAOM) framework is one of the most popular such approaches in the behavioral sciences, and is a particularly natural way to represent systems in which tie formation and maintenance depend on characteristics of some set of potentially interconnected nodes, and where also the interconnections, once formed, can “feed back� and affect nodal characteristics. In Part 1 of this presentation, I will describe the basic structure of a SAOM as a set of interrelated continuous-time Markov processes, and show how transition probabilities can be specified as functions of linkage and nodal data. In Part 2, an empirical application will be given, where the goal was to detect indirect (“spillover�) effects on alcohol use of an intervention with a subset of heavy drinkers in a college freshmen class.
-
 Benjamin Bolduc
Benjamin BolducViruses of bacteria and archaea are likely to be critical to all natural, engineered and human ecosystems, and yet their study is hampered by the lack of a universal or scalable taxonomic framework. Here, we introduce vConTACT 2.0, a network-based application to establish prokaryotic virus taxonomy that scales to thousands of uncultivated virus genomes/fragments, and integrates confidence scores for all taxonomic predictions. Performance tests using vConTACT 2.0 demonstrate near-identical correspondence to the current official viral taxonomy (>85% genus-rank assignments at 96% accuracy) through an integrated distance-based hierarchical approach. Beyond “known viruses�, we used vConTACT 2.0 to automatically assign 1,364 previously unclassified reference viruses to tentative taxa, and scaled it to modern metagenomic datasets for which the reference network was robust to adding 16,000 viral metagenomic contigs. Together these efforts provide a systematic reference network and an accurate, scalable taxonomic analysis tool that is critically needed for the research community.
-
 Subhadeep Paul
Subhadeep PaulDetecting communities or a group of vertices that are similar to each other in a complex network is a well-studied problem in network science. Methods for community detection include model-based approaches, namely, based on the stochastic block model and its variants, latent space models, as well as model-free approaches including spectral, modularity and matrix factorization methods. The stochastic block model is the most commonly employed model of complex networks with community structure. However, the model fails to explain many observed properties of networks including, heterogeneous and power-law degree distribution (scale-free), strong local clustering especially triadic closures (transitivity) in an otherwise sparse graph, hierarchical nature of connections (core-periphery structure). The goals of this project are to develop models for networks with community structure that explain observed properties of real-world networks better. We propose a superimposed stochastic block model (SupSBM) and a hyperbolic latent space network community model (Hypcomm) for this purpose. We analyze the performance of a higher-order spectral clustering algorithm under the SupSBM. We also develop computationally efficient Variational EM, and Laplacian spectral embedding algorithms to estimate the latent positions and the communities in the Hypcomm model.
-
 Facundo Memoli
Facundo MemoliWhen studying flocking/swarming behaviors in animals one is interested in quantifying and comparing the dynamics of the clustering induced by the coalescence and disbanding of animals in different groups. In a similar vein, studying the dynamics of social networks leads to the problem of characterizing groups/communities as they form and disperse throughout time.
Motivated by this, we study the problem of obtaining persistent homology based summaries of time-dependent data. Given a finite dynamic graph (DG), we first construct a zigzag persistence module arising from linearizing the dynamic transitive graph naturally induced from the input DG. Based on standard results, we then obtain a persistence diagram or barcode from this zigzag persistence module. We prove that these barcodes are stable under perturbations in the input DG under a suitable distance between DGs that we identify.
More precisely, our stability theorem can be interpreted as providing a lower bound for the distance between DGs. Since it relies on barcodes, and their bottleneck distance, this lower bound can be computed in polynomial time from the DG inputs.
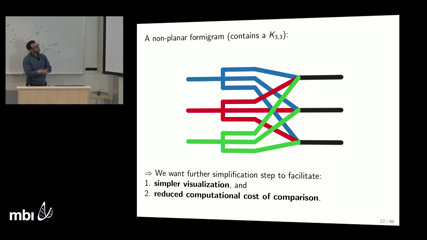
Along the way, we propose a summarization of dynamic graphs that captures their time-dependent clustering features which we call formigrams. These set-valued functions generalize the notion of dendrogram, a prevalent tool for hierarchical clustering. In order to elucidate the relationship between our distance between two DGs and the bottleneck distance between their associated barcodes, we exploit recent advances in the stability of zigzag persistence due to Botnan and Lesnick, and to Bjerkevik.
This is joint work with Woojin Kim.
https://research.math.osu.edu/networks/formigrams/
-
 Jennifer Hellmann
Jennifer HellmannIn group-living species, natural selection should favor behavioral strategies that collectively give rise to structures that facilitate group persistence and minimize conflict among group members. However, the extent to which conflict is present in a group, as well as how conflict is resolved, varies widely among groups; some groups have relatively little conflict while others disband due to high levels of conflict. Here, we seek to understand how both within-group and between-group changes alter group stability and social network structure in the group-living cichlid, Neolamprologus pulcher. We find that network changes can be related to both within-group and between-group differences in social structure. First, we demonstrate that there are unique characteristics of groups that remain intact compared to those that disband. Second, we demonstrate that group characteristics and the presence of neighbors alter the target of conflict within the group, shifting how aggressive, submissive, and affiliative interactions are divided among individuals of different sizes and dominance statuses. Collectively, our results demonstrate that both within and between group social structure should play a key role in predicting conflict within groups and group persistence.
-
 Kezia Manlove
Kezia ManloveUnderstanding dynamics of social contact is a key precursor for modeling propagation of pathogens, genes, and information through societies. However, there is little consistency within the ecological community about what form of social contact network to use, when. In this talk, I'll propose a tripartite network model borrowing from Lagrangian descriptions of animal movement, that relates disparate ecological networks under a single unifying structure using nodes of type Individual, Location, and Time. I'll show how this tripartite network can be reduced to a variety of different commonly used ecological networks, including individual association networks, home-range overlap networks, and transportation-like networks; and then I'll argue that particular social structures generate consistent redundancies in information between these node types. I'll then demonstrate how information derived from alternative ecological metrics like group size or home range size distributions can be used to constrain the tripartite network's topology. Lastly, I'll outline two potential pathways for using the tripartite network to formally quantify information lost through network projection, one reliant on graph theory, and the other reliant on Shannon's information.
