-
Duan Chen
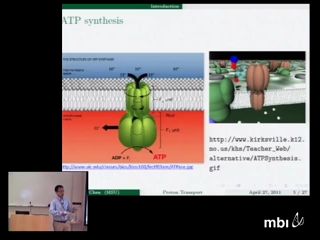
Proton transport across membranes is one of the most important and interesting phenomena in living cells. The present work proposes a multiscale/multiphysical model for the understanding of atomic level mechanism of proton transport in transmembrane proteins. We describe proton dynamics quantum mechanically via a density functional approach while implicitly model numerous solvent molecules as a dielectric continuum to reduce the number of degrees of freedom. The impact of protein molecular structure and its charge polarization on the proton transport is considered explicitly in atomic level. The molecular surface of the channel protein is utilized to split the discrete protein domain and the continuum solvent domain, and facilitate the multiscale discrete/continuum/quantum descriptions. We formulate a total free energy functional to put proton kinetic and potential energies as well as electrostatic energy of all ions on an equal footing. Generalized Poisson-Boltzmann equation and Kohn-Sham equation are obtained from the variational framework. A number of mathematical algorithms, including the Dirichlet to Neumann mapping, matched interface and boundary method, Gummel iteration, and Krylov space techniques are utilized to implement the proposed model in a computationally efficient manner. The Gramicidin A (GA) channel is used to demonstrate the performance of the proposed proton channel model and validate the efficiency of the proposed mathematical algorithms. The electrostatic characteristics and proton conductance of the GA channel are analyzed with a wide range of model parameters. A comparison with experimental data verifies the present model predictions and validates the proposed model.
-

Joseph Jerome
The Navier-Stokes/Poisson-Nernst-Planck model assumes significance because of its connection to the electrophysiology of the cell, including neuronal cell monitoring and microfluidic devices in biochip technology. The model has also been used in other applications, including electro- osmosis. The steady model is especially important in ion channel model- ing, because the channel remains open for milliseconds, and the transients appear to decay on the scale of tens of nanoseconds. In this talk, empha- sis will be placed upon the mathematical consistency of the unsteady and steady models. Some representative applications and simulations will be included.
-
Dirk Gillespie
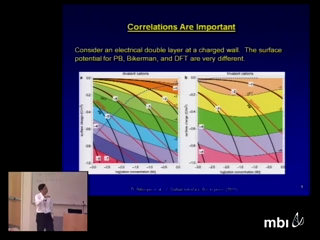
Theories like Poisson-Nernst-Planck that model ions as point charge are very useful in many applications. However, when ions are near highly-charged binding sites on proteins or inside ion channels, the size of the ions produces first-order effects because the ions' concentration is very large and/or because the ions are in a crevice or pore that is not much wider than the ions themselves. Density Functional Theory (DFT) of electrolytes (not electron orbitals) is a thermodynamically-derived theory that includes the effect of ion size in confining geometries. Applications of DFT to be discussed are as varied as modeling of ions at dielectric interfaces and ion currents through the ryanodine receptor calcium channel and through nanofluidic devices.
-
Qiong Zheng
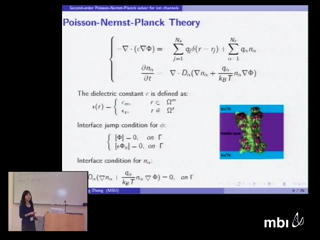
As a mean-field continuum model, Poisson Nernst-Planck (PNP) theory is an efficient computational tool for the study of ion transport phenomenon in the biological systems such as ion channels, which are important in the cell survival and the regulation of cellular activity. The present talk reports advanced numerical schemes and modified PNP models for ion channels. Based on our matched interface and boundary (MIB) method, we constructs second order convergent numerical scheme to efficiently solve the PNP equations in the presence of realistic macromolecular geometries and singular charges. Numerical applications are carried out to the Gramicidin A (GA) channel protein. Good agreement between our theoretical prediction and experimental measurements is found over a wide range of external voltages and concentrations. We also develop two modified PNP models to achieve either better computational efficiency or better prediction accuracy. One of our models serves as a simplified description of a multiple ion species system at the presence of external voltages, and the other incorporates the anisotropic property of certain biomolecular systems in inhomogeneous media.
-

Robert Eisenberg
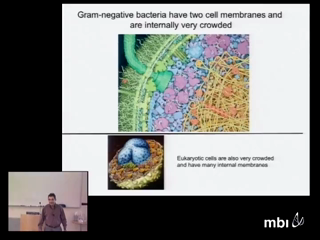
Ion channels are irresistible objects for biological study because they are the 'nanovalves of life' controlling most biological functions, much as transistors control computers. Channels contain an enormous density of crowded charged spheres, fixed and mobile, and induced polarization charge as well. Direct simulation of channel behavior in atomic detail is difficult if not impossible. Gaps in scales of time, volume, and concentration between atoms and biological systems are each ~1012. All the gaps must be dealt with at once, because biology deals with all the scales at once.
Simple models are surprisingly successful in dealing with ion binding in two very different (and important) channels over a large range of conditions, suggesting that mathematical analysis is both possible and useful. Amazingly, the same model with the same two parameters accounts quantitatively for qualitatively different binding in a wide range conditions for the very different calcium and sodium channels. The binding free energy is an output of the calculation, produced by the crowding of charged spheres in a very small space. The model does not involve any traditional chemical 'quantum' binding energies at all.
How can such a simple model give such specific results when crystallographic wisdom and chemical intuition says that selectivity depends on the precise structural relation of ions and side chains? The answer is that structure is the computed consequence of the forces in this model and is very important, but as an output of the model, not as an input. The relationship of ions and protein side chains changes when almost any condition is changed. Binding is a consequence of the 'induced fit' of side chains to ions and ions to side chains. Binding sites are self-organized and at their free energy minimum, forming different structures in different conditions.
Channels function away from equilibrium. A variational approach is obviously needed to replace our equilibrium analysis of binding and one is well under way, applying the energy variational methods previously perfected for more complex systems in electro-rheology by Chun Liu, his associates Rolf Ryham, and Yunkyong Hyon, and their colleague Yoichiro Mori.
-

Zhijun Wu
Structural properties on protein residue-level, such as the distances between two residues and the angles formed by short sequences of residues, can be important for structural analysis and modeling, but they have not been examined and documented in great detail. While these properties are difficult to measure experimentally, they can be statistically estimated based on their distributions in known protein structures.
Our work has involved in developing databases and software packages to estimate, document, and analyze the statistical distributions and correlations of various residue-level protein structural properties. We have found the high probability distributions of these properties and strong correlations among some of them. The results provide systematic and quantitative assessments on these properties, which can otherwise be estimated only individually and qualitatively.
Of particular interest is our recent work on an R-package called PRESS for the study of protein residue-level structural statistics. With this software, we can compute and display statistical distributions and correlations of certain residue-level structural properties in known protein structures, and use them to define statistical potentials and generate residue-level Ramachandran-like plots for structural analysis and assessment.
-- Joint work with Yuanyuan Huang, Stephen Bonett, Andrzej Kloczkowiski, and Robert Jernigan.
-

Ridgway Scott
We show how mathematics can help in the complex process of drug discovery. We give an example of modification of a common cancer drug that reduces unwanted side effects. The mathematical model used to do this relates to the hydrophobic effect, something not yet fully understood. The hydrophobic effect modulates the dielectric behavior of water, and this has dramatic effects on how we process drugs. Future mathematical advances in this area promise to make drug discovery more rational, and thus more rapid and predictable, and less costly.
-

Emil Alexov
Human DNA sequence differs among individuals and the most common variations are known as single nucleotide polymorphisms, or SNPs. Studies have shown that non-synonymous coding SNPs (nsSNPs - SNPs occurring in protein coding regions which lead to amino acid substitutions) can be responsible for many human diseases or cause the natural differences among the individuals by affecting the structure, function, interactions and other properties of expressed proteins. Of particular interest for us are rare missense mutations causing mental disorders by affecting the wild type characteristics of a certain protein. In this talk we will focus on three cases, spermine synthase, CLIC2 and SLC8A6 proteins, missense mutations in which were clinically shown to cause mental disorders. We demonstrate that in vast majority of the cases the mutations do not directly affect the functional properties of the corresponding protein, but rather indirectly alter its wild type characteristics. Further we contrast the effects caused by disease-causing missense mutations and naturally occurring harmless nsSNPs. It is demonstrated that disease-causing mutations do not necessary destabilize protein stability or protein-protein interactions, but can be stabilizing and still be harmful. Overall, a detailed computational analysis combined with an analysis of the corresponding biological function is needed to make reasonable prediction of the nature of the missense mutation.
-
Shan Zhao
Recently, we have introduced a differential geometry based model, the minimal molecular surface, to characterize the dielectric boundary between biomolecules and the surrounding aqueous environment. The mean curvature flow is used to minimize a surface free energy functional to drive the surface formation and evolution. More recently, several potential driven geometric flow models have been introduced in the literature for the analysis and computation of the equilibrium property of solvation, by appropriately coupling polar and nonpolar contributions in the free energy functional. The solvent-solute interface is usually treated as a sharp interface with discontinuous dielectric profile in a Lagrangian formulation, while in an Eulerian formulation a smeared interface model with continuous dielectric profile provides a convenient setting for solvation calculations. In the present study, we further extend the smeared interface model by considering a generalized nonlinear Poisson-Boltzmann (PB) equation in order to account for the salt effect. A new pseudo-time coupling between the surface geometric flows and electrostatic PB potential is introduced. Such a coupling allows for a fast numerical solution of governing nonlinear partial differential equations. Example solvation analysis of both small compounds and proteins are carried out to examine the proposed models and numerical approaches. Numerical results are compared to the experimental measurements and to those obtained by using other theoretical methods in the literature.
-

Nathan Baker
Implicit solvent models are important components of modern biomolecular simulation methodology due to their efficiency and dramatic reduction of dimensionality. However, such models are often constructed in an ad hoc manner with an arbitrary decomposition and specification of the polar and nonpolar components. In this talk, we review current implicit solvent models and suggest a new free energy functional which combines both polar and nonpolar solvation terms in a common self-consistent framework. Upon variation, this new free energy functional yields the traditional Poisson-Boltzmann equation as well as a new geometric flow equation. We describe numerical methods for solving these equations and comment on future research directions in this area.
This work was performed in collaboration with Dennis Thomas, Jaehun Chun, Zhan Chen, and Guowei Wei.
-

Christine Heitsch
Understanding how biological sequences encode structural and functional information is a fundamental scientific challenge. For RNA viral genomes, the information encoded in the sequence extends well-beyond their protein coding role to the role of intra-sequence base pairing in viral packaging, replication, and gene expression. Working with the Pariacoto virus as a model sequence, we investigate the compatibility of predicted base pairings with the dodecahedral cage known from crystallographic studies.
To build a putative secondary structure, we first analyze different possible configurations using a combinatorial model of RNA folding.
We give results on the trade-offs among types of loop structures, the asymptotic degree of branching in typical configurations, and the characteristics of stems in "well-determined" substructures. These mathematical results yield insights into the interaction of local and global constraints in RNA secondary structures, and suggest new directions in understanding the folding of RNA viral genomes.
-

Julie Mitchell
We present a computationally efficient method for flexible refinement of docking predictions that reflects observed motions within a protein's structural class. Using structural homologs, we derive deformation models that capture likely motions. The models or "replicates" typically align along a rigid core, with a handful of flexible loops, linkers and tails.
A few replicates can generate a much larger number of conformers, by exchanging each flexible region independently of the others. In this way, 10 replicates of a protein having 6 flexible regions can be used to generate a million conformations of a molecule. While this has obvious advantages in terms of sampling, the cost of assessing energies at every conformer is prohibitive, particularly when both molecules are flexible. Our approach addresses this combinatorial explosion, using key assumptions to compress the sampling by many orders of magnitude.
ReplicOpter can perform hierarchical clustering from a list of rigid docking predictions and find nearby structures to any promising cluster representatives. These predicted complexes can then be refined and rescored. ReplicOpter's scoring function includes a Lennard-Jones potential softened using the Anderson-Chandler-Weeks decomposition, a desolvation term derived from the Atomic Contact Energy function, Coulombic electrostatics, hydrogen bonding, and terms to model pi-pi and pi-cation interactions.
ReplicOpter has performed well on several recent CAPRI systems. We are presently benchmarking ReplicOpter on the complete docking benchmark set to fully establish its utility in refining rigid docking predictions and identifying near-native solutions.
-
Lukas Tamm
Structures of membrane proteins have been challenging to solve by any structural technique. We are developing solution NMR spectroscopy as a tool to study the structure and dynamics of membrane proteins, including bacterial outer membrane porins. This class of membrane proteins has proven particularly beneficial for these studies because (i) a larger chemical shift dispersion of residues is observed in β-sheets than in α-helices and (ii) much larger numbers of long-range NOEs can be observed in β-sheet vs. α-helical membrane proteins. Progress in this area will be illustrated with the small ion pore OmpA [1]. The gating of the OmpA ion channel has been studied by electrophysiological and thermodynamic approaches [2] and attempts to correlate these findings with dynamical properties of the protein will be illustrated [3]. Structural refinements can be obtained by including residual dipolar couplings and paramagnetic relaxation enhancements [4]. The methods have also been extended to solving the solution structure of the 33 kDa pH-gated porin OmpG embedded in a protein/DPC complex estimated to be about 80-90 kDa [5].
References:
1. Arora, A., Abildgaard, F., Bushweller, J.H., and Tamm, L.K. (2001) Structure of the outer membrane protein A transmembrane domain in detergent micelles by NMR spectroscopy. Nature Struct. Biol. 8:334-338.
2. Hong, H., Szabo, G., and Tamm, L.K. (2006) Electrostatic side-chain couplings in the gating of the OmpA ion channel suggest a mechanism for pore opening. Nature Chem. Biol. 11:627-635
3. Liang, B., Arora, A., and Tamm, L.K. (2010) Fast-time scale dynamics of outer membrane protein A by extended model-free analysis of NMR relaxation data. (Special issue: dynamics of membrane proteins by NMR) Biochim. Biophys. Acta 1798:68-76.
4. Liang, B., Bushweller, J.H., and Tamm, L.K. (2006) Site-directed parallel spin-labeling and paramagnetic relaxation enhancement in structure determination of membrane proteins by solution NMR. J.A.C.S., 126:4389-439.
5. Liang, B. and Tamm, L.K. (2007) Structure of outer membrane protein G by solution NMR. PNAS 104:16140-1614
-

Jack Quine
Differential geometry of curves uses the Frenet-Serret moving frame. A curve can be defined by scalar quantities of curvature and torsion and these quantities are defined by differentiating the frame. Similar techniques can be used for discrete curves formed by sequences of bonded atoms. The frames are related to molecular frames and are useful in finding protein structure from NMR data which gives the orientation of the frames with respect to the magnetic field direction. Curvature and torsion can be related to helical protein secondary structure. We discuss the use of these techniques in solving an alpha helical structure using NMR.
-

Ron Larson
We utilize the MARTINI coarse-grained force field to simulate lipid monolayers during the compression and re-expansion, to determine the effect of monolayer components on lung surfactant functioning. Our simulated monolayers contain pure dipalmitoylphosphatidylcholine (DPPC) and DPPC mixed with palmitoyloleoylphosphatidylglycerol (POPG), palmitic acid (PA), and/or peptides. The peptides considered include the 25-residue N-terminal fragment of SP-B (SP-B1-25), SP-C, and several SP-B1-25 mutants in which charged and hydrophilic residues are replaced by hydrophobic ones, or vice-versa. We observe two folding mechanisms: folding by the amplification of undulations and folding by nucleation about a defect. The first mechanism is observed in monolayers containing either POPG or peptides, while the second mechanism is observed only with peptides present, and involves the lipid-mediated aggregation of the peptides into a defect, from which the fold can nucleate. Fold nucleation from a defect displays a dependence on the hydrophobic character of the peptides; if the number of hydrophobic residues is decreased significantly, monolayer folding does not occur. The addition of POPG or peptides to the DPPC monolayer has a fluidizing effect, which assists monolayer folding. In contrast, the addition of PA has a charge-dependent condensing affect on DPPC monolayers containing SP-C. The peptides appear to play a significant role in the folding process, and provide a larger driving force for folding than POPG. In addition to promoting fold formation, the peptides also display fusogenic behavior, which can lead to surface refining.
-
Benzhuo Lu
Continuum modeling can be a proper choice to overcome the limitations on time and length scales of all-atom biomolecular simulations. The main concerns in this area are the model's accuracy and the numerical techniques/implementation. Besides, the molecular surface/volume meshing is also an unavoidable issue in many cases. I'll talk about our works on calculations of the Poisson-Boltzmann electrostatics, electro-diffusion-reaction simulations described by the Poisson-Nernst-Planck equations, and a general size-modified PNP/PB model. The latter method is applied to both equilibrium electrostatic calculation and rate prediction for diffusion-controlled enzyme-substrate reaction, and the particle (either ions or substrate molecule) size-effects to their concentrations and the reaction rate will be reported. Finally, I'll talk about our recently developed method and software TMSmesh for molecular surface meshing, which seems to be a useful tool to enable future macromolecular simulation using boundary element, finite element, or some other numerical methods.
 Duan ChenProton transport across membranes is one of the most important and interesting phenomena in living cells. The present work proposes a multiscale/multiphysical model for the understanding of atomic level mechanism of proton transport in transmembrane proteins. We describe proton dynamics quantum mechanically via a density functional approach while implicitly model numerous solvent molecules as a dielectric continuum to reduce the number of degrees of freedom. The impact of protein molecular structure and its charge polarization on the proton transport is considered explicitly in atomic level. The molecular surface of the channel protein is utilized to split the discrete protein domain and the continuum solvent domain, and facilitate the multiscale discrete/continuum/quantum descriptions. We formulate a total free energy functional to put proton kinetic and potential energies as well as electrostatic energy of all ions on an equal footing. Generalized Poisson-Boltzmann equation and Kohn-Sham equation are obtained from the variational framework. A number of mathematical algorithms, including the Dirichlet to Neumann mapping, matched interface and boundary method, Gummel iteration, and Krylov space techniques are utilized to implement the proposed model in a computationally efficient manner. The Gramicidin A (GA) channel is used to demonstrate the performance of the proposed proton channel model and validate the efficiency of the proposed mathematical algorithms. The electrostatic characteristics and proton conductance of the GA channel are analyzed with a wide range of model parameters. A comparison with experimental data verifies the present model predictions and validates the proposed model.
Duan ChenProton transport across membranes is one of the most important and interesting phenomena in living cells. The present work proposes a multiscale/multiphysical model for the understanding of atomic level mechanism of proton transport in transmembrane proteins. We describe proton dynamics quantum mechanically via a density functional approach while implicitly model numerous solvent molecules as a dielectric continuum to reduce the number of degrees of freedom. The impact of protein molecular structure and its charge polarization on the proton transport is considered explicitly in atomic level. The molecular surface of the channel protein is utilized to split the discrete protein domain and the continuum solvent domain, and facilitate the multiscale discrete/continuum/quantum descriptions. We formulate a total free energy functional to put proton kinetic and potential energies as well as electrostatic energy of all ions on an equal footing. Generalized Poisson-Boltzmann equation and Kohn-Sham equation are obtained from the variational framework. A number of mathematical algorithms, including the Dirichlet to Neumann mapping, matched interface and boundary method, Gummel iteration, and Krylov space techniques are utilized to implement the proposed model in a computationally efficient manner. The Gramicidin A (GA) channel is used to demonstrate the performance of the proposed proton channel model and validate the efficiency of the proposed mathematical algorithms. The electrostatic characteristics and proton conductance of the GA channel are analyzed with a wide range of model parameters. A comparison with experimental data verifies the present model predictions and validates the proposed model. Joseph JeromeThe Navier-Stokes/Poisson-Nernst-Planck model assumes significance because of its connection to the electrophysiology of the cell, including neuronal cell monitoring and microfluidic devices in biochip technology. The model has also been used in other applications, including electro- osmosis. The steady model is especially important in ion channel model- ing, because the channel remains open for milliseconds, and the transients appear to decay on the scale of tens of nanoseconds. In this talk, empha- sis will be placed upon the mathematical consistency of the unsteady and steady models. Some representative applications and simulations will be included.
Joseph JeromeThe Navier-Stokes/Poisson-Nernst-Planck model assumes significance because of its connection to the electrophysiology of the cell, including neuronal cell monitoring and microfluidic devices in biochip technology. The model has also been used in other applications, including electro- osmosis. The steady model is especially important in ion channel model- ing, because the channel remains open for milliseconds, and the transients appear to decay on the scale of tens of nanoseconds. In this talk, empha- sis will be placed upon the mathematical consistency of the unsteady and steady models. Some representative applications and simulations will be included. Dirk GillespieTheories like Poisson-Nernst-Planck that model ions as point charge are very useful in many applications. However, when ions are near highly-charged binding sites on proteins or inside ion channels, the size of the ions produces first-order effects because the ions' concentration is very large and/or because the ions are in a crevice or pore that is not much wider than the ions themselves. Density Functional Theory (DFT) of electrolytes (not electron orbitals) is a thermodynamically-derived theory that includes the effect of ion size in confining geometries. Applications of DFT to be discussed are as varied as modeling of ions at dielectric interfaces and ion currents through the ryanodine receptor calcium channel and through nanofluidic devices.
Dirk GillespieTheories like Poisson-Nernst-Planck that model ions as point charge are very useful in many applications. However, when ions are near highly-charged binding sites on proteins or inside ion channels, the size of the ions produces first-order effects because the ions' concentration is very large and/or because the ions are in a crevice or pore that is not much wider than the ions themselves. Density Functional Theory (DFT) of electrolytes (not electron orbitals) is a thermodynamically-derived theory that includes the effect of ion size in confining geometries. Applications of DFT to be discussed are as varied as modeling of ions at dielectric interfaces and ion currents through the ryanodine receptor calcium channel and through nanofluidic devices. Qiong ZhengAs a mean-field continuum model, Poisson Nernst-Planck (PNP) theory is an efficient computational tool for the study of ion transport phenomenon in the biological systems such as ion channels, which are important in the cell survival and the regulation of cellular activity. The present talk reports advanced numerical schemes and modified PNP models for ion channels. Based on our matched interface and boundary (MIB) method, we constructs second order convergent numerical scheme to efficiently solve the PNP equations in the presence of realistic macromolecular geometries and singular charges. Numerical applications are carried out to the Gramicidin A (GA) channel protein. Good agreement between our theoretical prediction and experimental measurements is found over a wide range of external voltages and concentrations. We also develop two modified PNP models to achieve either better computational efficiency or better prediction accuracy. One of our models serves as a simplified description of a multiple ion species system at the presence of external voltages, and the other incorporates the anisotropic property of certain biomolecular systems in inhomogeneous media.
Qiong ZhengAs a mean-field continuum model, Poisson Nernst-Planck (PNP) theory is an efficient computational tool for the study of ion transport phenomenon in the biological systems such as ion channels, which are important in the cell survival and the regulation of cellular activity. The present talk reports advanced numerical schemes and modified PNP models for ion channels. Based on our matched interface and boundary (MIB) method, we constructs second order convergent numerical scheme to efficiently solve the PNP equations in the presence of realistic macromolecular geometries and singular charges. Numerical applications are carried out to the Gramicidin A (GA) channel protein. Good agreement between our theoretical prediction and experimental measurements is found over a wide range of external voltages and concentrations. We also develop two modified PNP models to achieve either better computational efficiency or better prediction accuracy. One of our models serves as a simplified description of a multiple ion species system at the presence of external voltages, and the other incorporates the anisotropic property of certain biomolecular systems in inhomogeneous media. Robert EisenbergIon channels are irresistible objects for biological study because they are the 'nanovalves of life' controlling most biological functions, much as transistors control computers. Channels contain an enormous density of crowded charged spheres, fixed and mobile, and induced polarization charge as well. Direct simulation of channel behavior in atomic detail is difficult if not impossible. Gaps in scales of time, volume, and concentration between atoms and biological systems are each ~1012. All the gaps must be dealt with at once, because biology deals with all the scales at once.
Robert EisenbergIon channels are irresistible objects for biological study because they are the 'nanovalves of life' controlling most biological functions, much as transistors control computers. Channels contain an enormous density of crowded charged spheres, fixed and mobile, and induced polarization charge as well. Direct simulation of channel behavior in atomic detail is difficult if not impossible. Gaps in scales of time, volume, and concentration between atoms and biological systems are each ~1012. All the gaps must be dealt with at once, because biology deals with all the scales at once. Zhijun WuStructural properties on protein residue-level, such as the distances between two residues and the angles formed by short sequences of residues, can be important for structural analysis and modeling, but they have not been examined and documented in great detail. While these properties are difficult to measure experimentally, they can be statistically estimated based on their distributions in known protein structures.
Zhijun WuStructural properties on protein residue-level, such as the distances between two residues and the angles formed by short sequences of residues, can be important for structural analysis and modeling, but they have not been examined and documented in great detail. While these properties are difficult to measure experimentally, they can be statistically estimated based on their distributions in known protein structures. Ridgway ScottWe show how mathematics can help in the complex process of drug discovery. We give an example of modification of a common cancer drug that reduces unwanted side effects. The mathematical model used to do this relates to the hydrophobic effect, something not yet fully understood. The hydrophobic effect modulates the dielectric behavior of water, and this has dramatic effects on how we process drugs. Future mathematical advances in this area promise to make drug discovery more rational, and thus more rapid and predictable, and less costly.
Ridgway ScottWe show how mathematics can help in the complex process of drug discovery. We give an example of modification of a common cancer drug that reduces unwanted side effects. The mathematical model used to do this relates to the hydrophobic effect, something not yet fully understood. The hydrophobic effect modulates the dielectric behavior of water, and this has dramatic effects on how we process drugs. Future mathematical advances in this area promise to make drug discovery more rational, and thus more rapid and predictable, and less costly. Emil AlexovHuman DNA sequence differs among individuals and the most common variations are known as single nucleotide polymorphisms, or SNPs. Studies have shown that non-synonymous coding SNPs (nsSNPs - SNPs occurring in protein coding regions which lead to amino acid substitutions) can be responsible for many human diseases or cause the natural differences among the individuals by affecting the structure, function, interactions and other properties of expressed proteins. Of particular interest for us are rare missense mutations causing mental disorders by affecting the wild type characteristics of a certain protein. In this talk we will focus on three cases, spermine synthase, CLIC2 and SLC8A6 proteins, missense mutations in which were clinically shown to cause mental disorders. We demonstrate that in vast majority of the cases the mutations do not directly affect the functional properties of the corresponding protein, but rather indirectly alter its wild type characteristics. Further we contrast the effects caused by disease-causing missense mutations and naturally occurring harmless nsSNPs. It is demonstrated that disease-causing mutations do not necessary destabilize protein stability or protein-protein interactions, but can be stabilizing and still be harmful. Overall, a detailed computational analysis combined with an analysis of the corresponding biological function is needed to make reasonable prediction of the nature of the missense mutation.
Emil AlexovHuman DNA sequence differs among individuals and the most common variations are known as single nucleotide polymorphisms, or SNPs. Studies have shown that non-synonymous coding SNPs (nsSNPs - SNPs occurring in protein coding regions which lead to amino acid substitutions) can be responsible for many human diseases or cause the natural differences among the individuals by affecting the structure, function, interactions and other properties of expressed proteins. Of particular interest for us are rare missense mutations causing mental disorders by affecting the wild type characteristics of a certain protein. In this talk we will focus on three cases, spermine synthase, CLIC2 and SLC8A6 proteins, missense mutations in which were clinically shown to cause mental disorders. We demonstrate that in vast majority of the cases the mutations do not directly affect the functional properties of the corresponding protein, but rather indirectly alter its wild type characteristics. Further we contrast the effects caused by disease-causing missense mutations and naturally occurring harmless nsSNPs. It is demonstrated that disease-causing mutations do not necessary destabilize protein stability or protein-protein interactions, but can be stabilizing and still be harmful. Overall, a detailed computational analysis combined with an analysis of the corresponding biological function is needed to make reasonable prediction of the nature of the missense mutation. Shan ZhaoRecently, we have introduced a differential geometry based model, the minimal molecular surface, to characterize the dielectric boundary between biomolecules and the surrounding aqueous environment. The mean curvature flow is used to minimize a surface free energy functional to drive the surface formation and evolution. More recently, several potential driven geometric flow models have been introduced in the literature for the analysis and computation of the equilibrium property of solvation, by appropriately coupling polar and nonpolar contributions in the free energy functional. The solvent-solute interface is usually treated as a sharp interface with discontinuous dielectric profile in a Lagrangian formulation, while in an Eulerian formulation a smeared interface model with continuous dielectric profile provides a convenient setting for solvation calculations. In the present study, we further extend the smeared interface model by considering a generalized nonlinear Poisson-Boltzmann (PB) equation in order to account for the salt effect. A new pseudo-time coupling between the surface geometric flows and electrostatic PB potential is introduced. Such a coupling allows for a fast numerical solution of governing nonlinear partial differential equations. Example solvation analysis of both small compounds and proteins are carried out to examine the proposed models and numerical approaches. Numerical results are compared to the experimental measurements and to those obtained by using other theoretical methods in the literature.
Shan ZhaoRecently, we have introduced a differential geometry based model, the minimal molecular surface, to characterize the dielectric boundary between biomolecules and the surrounding aqueous environment. The mean curvature flow is used to minimize a surface free energy functional to drive the surface formation and evolution. More recently, several potential driven geometric flow models have been introduced in the literature for the analysis and computation of the equilibrium property of solvation, by appropriately coupling polar and nonpolar contributions in the free energy functional. The solvent-solute interface is usually treated as a sharp interface with discontinuous dielectric profile in a Lagrangian formulation, while in an Eulerian formulation a smeared interface model with continuous dielectric profile provides a convenient setting for solvation calculations. In the present study, we further extend the smeared interface model by considering a generalized nonlinear Poisson-Boltzmann (PB) equation in order to account for the salt effect. A new pseudo-time coupling between the surface geometric flows and electrostatic PB potential is introduced. Such a coupling allows for a fast numerical solution of governing nonlinear partial differential equations. Example solvation analysis of both small compounds and proteins are carried out to examine the proposed models and numerical approaches. Numerical results are compared to the experimental measurements and to those obtained by using other theoretical methods in the literature. Nathan BakerImplicit solvent models are important components of modern biomolecular simulation methodology due to their efficiency and dramatic reduction of dimensionality. However, such models are often constructed in an ad hoc manner with an arbitrary decomposition and specification of the polar and nonpolar components. In this talk, we review current implicit solvent models and suggest a new free energy functional which combines both polar and nonpolar solvation terms in a common self-consistent framework. Upon variation, this new free energy functional yields the traditional Poisson-Boltzmann equation as well as a new geometric flow equation. We describe numerical methods for solving these equations and comment on future research directions in this area.
Nathan BakerImplicit solvent models are important components of modern biomolecular simulation methodology due to their efficiency and dramatic reduction of dimensionality. However, such models are often constructed in an ad hoc manner with an arbitrary decomposition and specification of the polar and nonpolar components. In this talk, we review current implicit solvent models and suggest a new free energy functional which combines both polar and nonpolar solvation terms in a common self-consistent framework. Upon variation, this new free energy functional yields the traditional Poisson-Boltzmann equation as well as a new geometric flow equation. We describe numerical methods for solving these equations and comment on future research directions in this area. Christine HeitschUnderstanding how biological sequences encode structural and functional information is a fundamental scientific challenge. For RNA viral genomes, the information encoded in the sequence extends well-beyond their protein coding role to the role of intra-sequence base pairing in viral packaging, replication, and gene expression. Working with the Pariacoto virus as a model sequence, we investigate the compatibility of predicted base pairings with the dodecahedral cage known from crystallographic studies.
Christine HeitschUnderstanding how biological sequences encode structural and functional information is a fundamental scientific challenge. For RNA viral genomes, the information encoded in the sequence extends well-beyond their protein coding role to the role of intra-sequence base pairing in viral packaging, replication, and gene expression. Working with the Pariacoto virus as a model sequence, we investigate the compatibility of predicted base pairings with the dodecahedral cage known from crystallographic studies. Julie MitchellWe present a computationally efficient method for flexible refinement of docking predictions that reflects observed motions within a protein's structural class. Using structural homologs, we derive deformation models that capture likely motions. The models or "replicates" typically align along a rigid core, with a handful of flexible loops, linkers and tails.
Julie MitchellWe present a computationally efficient method for flexible refinement of docking predictions that reflects observed motions within a protein's structural class. Using structural homologs, we derive deformation models that capture likely motions. The models or "replicates" typically align along a rigid core, with a handful of flexible loops, linkers and tails. Lukas TammStructures of membrane proteins have been challenging to solve by any structural technique. We are developing solution NMR spectroscopy as a tool to study the structure and dynamics of membrane proteins, including bacterial outer membrane porins. This class of membrane proteins has proven particularly beneficial for these studies because (i) a larger chemical shift dispersion of residues is observed in β-sheets than in α-helices and (ii) much larger numbers of long-range NOEs can be observed in β-sheet vs. α-helical membrane proteins. Progress in this area will be illustrated with the small ion pore OmpA [1]. The gating of the OmpA ion channel has been studied by electrophysiological and thermodynamic approaches [2] and attempts to correlate these findings with dynamical properties of the protein will be illustrated [3]. Structural refinements can be obtained by including residual dipolar couplings and paramagnetic relaxation enhancements [4]. The methods have also been extended to solving the solution structure of the 33 kDa pH-gated porin OmpG embedded in a protein/DPC complex estimated to be about 80-90 kDa [5].
Lukas TammStructures of membrane proteins have been challenging to solve by any structural technique. We are developing solution NMR spectroscopy as a tool to study the structure and dynamics of membrane proteins, including bacterial outer membrane porins. This class of membrane proteins has proven particularly beneficial for these studies because (i) a larger chemical shift dispersion of residues is observed in β-sheets than in α-helices and (ii) much larger numbers of long-range NOEs can be observed in β-sheet vs. α-helical membrane proteins. Progress in this area will be illustrated with the small ion pore OmpA [1]. The gating of the OmpA ion channel has been studied by electrophysiological and thermodynamic approaches [2] and attempts to correlate these findings with dynamical properties of the protein will be illustrated [3]. Structural refinements can be obtained by including residual dipolar couplings and paramagnetic relaxation enhancements [4]. The methods have also been extended to solving the solution structure of the 33 kDa pH-gated porin OmpG embedded in a protein/DPC complex estimated to be about 80-90 kDa [5]. Jack QuineDifferential geometry of curves uses the Frenet-Serret moving frame. A curve can be defined by scalar quantities of curvature and torsion and these quantities are defined by differentiating the frame. Similar techniques can be used for discrete curves formed by sequences of bonded atoms. The frames are related to molecular frames and are useful in finding protein structure from NMR data which gives the orientation of the frames with respect to the magnetic field direction. Curvature and torsion can be related to helical protein secondary structure. We discuss the use of these techniques in solving an alpha helical structure using NMR.
Jack QuineDifferential geometry of curves uses the Frenet-Serret moving frame. A curve can be defined by scalar quantities of curvature and torsion and these quantities are defined by differentiating the frame. Similar techniques can be used for discrete curves formed by sequences of bonded atoms. The frames are related to molecular frames and are useful in finding protein structure from NMR data which gives the orientation of the frames with respect to the magnetic field direction. Curvature and torsion can be related to helical protein secondary structure. We discuss the use of these techniques in solving an alpha helical structure using NMR. Ron LarsonWe utilize the MARTINI coarse-grained force field to simulate lipid monolayers during the compression and re-expansion, to determine the effect of monolayer components on lung surfactant functioning. Our simulated monolayers contain pure dipalmitoylphosphatidylcholine (DPPC) and DPPC mixed with palmitoyloleoylphosphatidylglycerol (POPG), palmitic acid (PA), and/or peptides. The peptides considered include the 25-residue N-terminal fragment of SP-B (SP-B1-25), SP-C, and several SP-B1-25 mutants in which charged and hydrophilic residues are replaced by hydrophobic ones, or vice-versa. We observe two folding mechanisms: folding by the amplification of undulations and folding by nucleation about a defect. The first mechanism is observed in monolayers containing either POPG or peptides, while the second mechanism is observed only with peptides present, and involves the lipid-mediated aggregation of the peptides into a defect, from which the fold can nucleate. Fold nucleation from a defect displays a dependence on the hydrophobic character of the peptides; if the number of hydrophobic residues is decreased significantly, monolayer folding does not occur. The addition of POPG or peptides to the DPPC monolayer has a fluidizing effect, which assists monolayer folding. In contrast, the addition of PA has a charge-dependent condensing affect on DPPC monolayers containing SP-C. The peptides appear to play a significant role in the folding process, and provide a larger driving force for folding than POPG. In addition to promoting fold formation, the peptides also display fusogenic behavior, which can lead to surface refining.
Ron LarsonWe utilize the MARTINI coarse-grained force field to simulate lipid monolayers during the compression and re-expansion, to determine the effect of monolayer components on lung surfactant functioning. Our simulated monolayers contain pure dipalmitoylphosphatidylcholine (DPPC) and DPPC mixed with palmitoyloleoylphosphatidylglycerol (POPG), palmitic acid (PA), and/or peptides. The peptides considered include the 25-residue N-terminal fragment of SP-B (SP-B1-25), SP-C, and several SP-B1-25 mutants in which charged and hydrophilic residues are replaced by hydrophobic ones, or vice-versa. We observe two folding mechanisms: folding by the amplification of undulations and folding by nucleation about a defect. The first mechanism is observed in monolayers containing either POPG or peptides, while the second mechanism is observed only with peptides present, and involves the lipid-mediated aggregation of the peptides into a defect, from which the fold can nucleate. Fold nucleation from a defect displays a dependence on the hydrophobic character of the peptides; if the number of hydrophobic residues is decreased significantly, monolayer folding does not occur. The addition of POPG or peptides to the DPPC monolayer has a fluidizing effect, which assists monolayer folding. In contrast, the addition of PA has a charge-dependent condensing affect on DPPC monolayers containing SP-C. The peptides appear to play a significant role in the folding process, and provide a larger driving force for folding than POPG. In addition to promoting fold formation, the peptides also display fusogenic behavior, which can lead to surface refining. Benzhuo LuContinuum modeling can be a proper choice to overcome the limitations on time and length scales of all-atom biomolecular simulations. The main concerns in this area are the model's accuracy and the numerical techniques/implementation. Besides, the molecular surface/volume meshing is also an unavoidable issue in many cases. I'll talk about our works on calculations of the Poisson-Boltzmann electrostatics, electro-diffusion-reaction simulations described by the Poisson-Nernst-Planck equations, and a general size-modified PNP/PB model. The latter method is applied to both equilibrium electrostatic calculation and rate prediction for diffusion-controlled enzyme-substrate reaction, and the particle (either ions or substrate molecule) size-effects to their concentrations and the reaction rate will be reported. Finally, I'll talk about our recently developed method and software TMSmesh for molecular surface meshing, which seems to be a useful tool to enable future macromolecular simulation using boundary element, finite element, or some other numerical methods.
Benzhuo LuContinuum modeling can be a proper choice to overcome the limitations on time and length scales of all-atom biomolecular simulations. The main concerns in this area are the model's accuracy and the numerical techniques/implementation. Besides, the molecular surface/volume meshing is also an unavoidable issue in many cases. I'll talk about our works on calculations of the Poisson-Boltzmann electrostatics, electro-diffusion-reaction simulations described by the Poisson-Nernst-Planck equations, and a general size-modified PNP/PB model. The latter method is applied to both equilibrium electrostatic calculation and rate prediction for diffusion-controlled enzyme-substrate reaction, and the particle (either ions or substrate molecule) size-effects to their concentrations and the reaction rate will be reported. Finally, I'll talk about our recently developed method and software TMSmesh for molecular surface meshing, which seems to be a useful tool to enable future macromolecular simulation using boundary element, finite element, or some other numerical methods.