-

Ryan Gutenkunst
Not available
-

Mark Siegal
Genetically identical cells grown in the same culture display striking cell-to-cell heterogeneity in gene expression and other traits. A crucial challenge is to understand how much of this heterogeneity reflects the noise tolerance of a robust system and how much serves a biological function. In some circumstances, heterogeneous traits might be favored over robust ones. For example, in bacteria cell-to-cell heterogeneity can serve as a bet-hedging mechanism, allowing a few cells to survive acute antibiotic stress while the others perish. Where a population of organisms falls on the continuum from uniformity to bet-hedging depends on the environmental regime it experiences. We describe a bet-hedging phenomenon in the yeast Saccharomyces cerevisiae, which occupies a range of natural and human-associated environments. We use a novel, high-throughput microscopy assay that monitors variable protein expression, growth rate and survival outcomes of tens of thousands of yeast microcolonies simultaneously. Clonal yeast populations display broad distributions of growth rates, and slow growth predicts resistance to acute heat stress. Expression of Tsl1, a trehalose-synthesis regulator, marks slow-growing cells and contributes to this resistance. I will present these results and discuss them in the context of the evolutionary forces shaping robust and heterogeneous traits.
-
Sayak Mukherjee
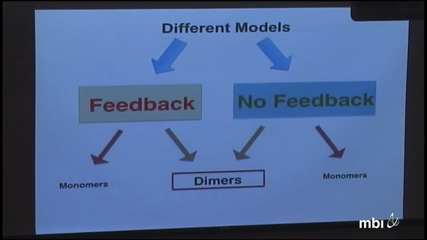
The process that warrants the generation of self tolerant peripheral T cells is called the thymocyte selection. During this maturation process, the overtly self reactive as well as unresponsive thymocytes are deleted from the cell population. The thymocytes equipped with T cell receptors (TCRs), capable of responding moderately to the self peptides are allowed to survive. Recently water soluble second messenger, inositol(1,3,4,5) tetrakisphosphate (IP4), has been implicated to play a crucial role in thymocyte positive selection (Huang et al.). It has been suggested that these IP4 molecules regulate the transient activation of the Tec- family protein tyrosine kinase Itk through a competing positive and negative feedbacks. The exact molecular mechanism involved in this feedback is however unclear. It is possible to con- struct more than one model with contrasting molecular mechanisms to explain the present body of experimental observations. This calls for criteria to choose among these models.
Robustness in face of the variation of the parameters in a model has been ubiquitously used as a criterion for model discrimination. Here we have used the maximum entropy, calculated with the constraints imposed by the experiments as a measure of robustness. Our data indicates that the models which are maximally robust share a cooperative allosteric mode of Itk regulation involving dimeric PH domains.
Joint work with Stephanie Rigaud, S. Seok, Agnieszka Prochenka, Guo Fu, Michael Dworkin, Nicholas R. J. Gascoigne, Veronika J. Vieland, Karsten Sauer, and Jayajit Das.
-
Ilya Nemenman
Ilya Nemenman, Physics, Emory University
Biochemical processes typically involve huge numbers of individual reversible steps, each with its own dynamical rate constants. For example, kinetic proofreading processes rely upon numerous sequential reactions in order to guarantee the precise construction of specific macromolecules. I will present a characterization of the first passage (completion) time distributions for such processes. I will argue that, for a wide class of biochemical kinetics systems related to kinetic proofreading, the completion time time behavior simplifies as the system size grows: it becomes either deterministic or exponentially distributed, with a very narrow transition between the two regimes. In both regimes, the dynamical complexity of the full system is trivial compared to its apparent structural complexity. This robust simplification of completion time distributions is independent of many microscopic details of the signaling systems and can be utilized for efficient control of cellular response properties. Even further simplifications are possible when one considers dynamics of many coupled kinetic proofreading enabled receptors, which can attain low activation noise with robust mean time to activation.
-

Maxim Artyomov
Screens monitoring the effects of deletion, knock-down or over expression of regulatory genes on the expression of their target genes are critical for deciphering the organization of complex regulatory networks. However, since perturbation assays cannot distinguish direct from indirect effects, the derived networks are significantly more complex than the true underlying one. Discovery of the true network organization is a long-standing challenge and several approaches have been developed to infer regulatory networks based on gene expression data. Recent studies indicate [ref] that information obtained from perturbation screens is critical for the ability to identify network structure accurately. This information is typically incorporated into network inference algorithms in two ways: first, the strength of the perturbation effect is translated into interaction confidence values and, second, topological analysis of the experimental networks is performed to find interactions that are most important to preserve network structure and, hence, are more likely to be biological. This underscores importance of having accurate methodology for accurate inference of network topology. In this work we present Exigo, an approach for systematic analysis of network topology. Exigo provides the means to identify core network structure for an input network of any topology with an arbitrary number of activating and inhibiting interactions. We further show that Exigo allows for significant improvement in the network inference. To illustrate this, we constructed a chimeric network inference method that incorporates Exigo into existing inference pipeline [ref], benchmarked it against DREAM challenge networks and found significant improvement in network inference compared to DREAM top performers.
-
Joanna Masel
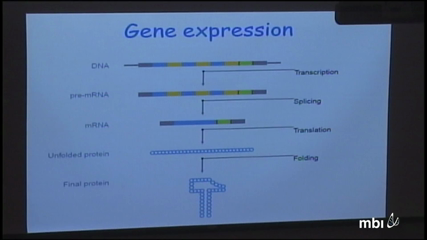
Making genes into gene products is subject to predictable errors, each with a phenotypic effect that depends on a normally cryptic sequence. The distribution of fitness effects of these cryptic sequences, like that of new mutations, is bimodal. For example, a cryptic sequence might be strongly deleterious if it causes protein misfolding, or it might have only a minor effect if it preserves the protein fold and tweaks function. Few sequences have effect sizes that fall in between.
Strongly deleterious sequences can be subject to some selection even while they are cryptic, and expressed only at low levels that depend on a molecular error. Robustness to the potentially deleterious effects of cryptic sequences can be achieved globally by avoiding making errors (e.g., via proofreading machinery) or locally by ensuring that each cryptic sequence has a relatively benign effect. The local solution requires powerful selection acting on every cryptic site, and so evolves only in large populations. Small populations with less effective selection evolve global proofreading solutions. However, we also find that for a large range of realistic intermediate population sizes, the evolutionary dynamics are bistable and either solution may result. The local solution, which does not occur in very small populations, facilitates the co-option of cryptic sequences and therefore substantially increases evolvability. This can occur even in genetically uniform populations, illustrating that neighbourhood richness or "quality" in genotype space can be more important to evolvability than the quantity of genotypes in a population spread across that space.
-

Jon Wilkins
When natural selection is acting at the level of the the individual phenotype, we expect selection to favor more robust phenotypes. However, selection acting at the level of the gene can undermine adaptation of the individual organism, and lead to the fixation of suboptimal traits. Genomic imprinting, the phenomenon where the pattern of expression of an allele depends on its parental origin, is thought to result from and evolutionary intragenomic conflict, where maternally and paternally inherited alleles favor different optimal phenotypes. Using a simple model, I will illustrate how this can lead to the systematic unravelling of phenotypic robustness.
-

Emilie Snell-Rood
Not available.
-

David Holloway
Fruit flies are models for understanding the genetic regulation involved in specifying the complex body plans of higher animals. The head-to-tail (anterior-posterior) axis of the fly (Drosophila) is established in the first hours of development. Maternally supplied factors form concentration gradients which direct embryonic (zygotic) genes where to be activated to express proteins. These protein patterns specify the positions and cell types of the body's tissues. Recent research has shown, comparing between embryos, that the zygotic gene products are much more precisely positioned than the maternal gradients, indicating an embryonic error reduction mechanism. Within embryos, there is the additional aspect that DNA and mRNA operate at very low copy number, and the associated high relative noise has the potential to strongly affect protein expression patterns. In recent work, we have focused on the noise aspects of positional specification within individual embryos, and what molecular mechanisms confer robustness to the process.
We simulate activation of hunchback (hb), a primary target of the maternal Bicoid (Bcd) protein gradient, which forms an expression pattern dividing the embryo into anterior and posterior halves. We use a master equation approach to simulate the stochastic dynamics of hb regulation, including the binding/unbinding of Bcd molecules at the hb promoter (the portion of DNA regulating hb transcription), hb transcription, subsequent translation to Hb protein, binding/unbinding of Hb at the promoter (self-regulation), and diffusion of the Bcd and Hb proteins. Model parameters were set by deterministically matching large scale pattern features for a series of experimental expression patterns: wild-type (WT) embryos; hb mutants lacking self-regulation; and constructs in which portions of the hb promoter were used to express a reporter gene (lacZ). The model was then solved stochastically to predict the noise output in these different experiments. In subsequent noise measurements we experimentally corroborated several predictions: including that mRNA is noisier than protein and that Hb self-regulation reduces noise.
Results indicate that WT (self-regulatory) Hb output noise is predominantly dependent on the transcription and translation dynamics of its own expression, and is uncorrelated with Bcd fluctuations. In the constructs and mutant, which lack self-regulation, we find that increasing the number and strength of Bcd binding sites (there are 6 in the core hb promoter) provides a rudimentary level of noise reduction. We have recently incorporated a known inhibitor of hb, Kr�ppel (Kr), into the model � preliminary indications are that the Hb-Kr activator-inhibitor dynamics increase precision at the mid-embryo boundary.
The analysis of hb shows common modes of gene regulation (e.g. multiple regulatory sites, self-regulation) that are involved in noise reduction, which can be applied generally to reproducibility and determinacy of spatial patterning in other developmental phenomena.
-

Ricardo Azevedo
Evolution is the movement of populations through a space of genotypes. This space can be modeled as an undirected network connecting genotypes that can be reached through mutation. In this view, the mutational robustness of a genotype is the proportion of its mutational neighbors that are viable. Robustness can facilitate the exploration of genotype networks, or evolvability. Sexual reproduction is also widely believed to promote evolvability, for two reasons. First, because it allows long jumps through genotype space. Second, because it selects for mutational robustness. Here, I show that, depending on the structure of the genotype network, sexual reproduction may not select for the highest mutational robustness, and can actually reduce evolvability.
 Ryan GutenkunstNot available
Ryan GutenkunstNot available Mark SiegalGenetically identical cells grown in the same culture display striking cell-to-cell heterogeneity in gene expression and other traits. A crucial challenge is to understand how much of this heterogeneity reflects the noise tolerance of a robust system and how much serves a biological function. In some circumstances, heterogeneous traits might be favored over robust ones. For example, in bacteria cell-to-cell heterogeneity can serve as a bet-hedging mechanism, allowing a few cells to survive acute antibiotic stress while the others perish. Where a population of organisms falls on the continuum from uniformity to bet-hedging depends on the environmental regime it experiences. We describe a bet-hedging phenomenon in the yeast Saccharomyces cerevisiae, which occupies a range of natural and human-associated environments. We use a novel, high-throughput microscopy assay that monitors variable protein expression, growth rate and survival outcomes of tens of thousands of yeast microcolonies simultaneously. Clonal yeast populations display broad distributions of growth rates, and slow growth predicts resistance to acute heat stress. Expression of Tsl1, a trehalose-synthesis regulator, marks slow-growing cells and contributes to this resistance. I will present these results and discuss them in the context of the evolutionary forces shaping robust and heterogeneous traits.
Mark SiegalGenetically identical cells grown in the same culture display striking cell-to-cell heterogeneity in gene expression and other traits. A crucial challenge is to understand how much of this heterogeneity reflects the noise tolerance of a robust system and how much serves a biological function. In some circumstances, heterogeneous traits might be favored over robust ones. For example, in bacteria cell-to-cell heterogeneity can serve as a bet-hedging mechanism, allowing a few cells to survive acute antibiotic stress while the others perish. Where a population of organisms falls on the continuum from uniformity to bet-hedging depends on the environmental regime it experiences. We describe a bet-hedging phenomenon in the yeast Saccharomyces cerevisiae, which occupies a range of natural and human-associated environments. We use a novel, high-throughput microscopy assay that monitors variable protein expression, growth rate and survival outcomes of tens of thousands of yeast microcolonies simultaneously. Clonal yeast populations display broad distributions of growth rates, and slow growth predicts resistance to acute heat stress. Expression of Tsl1, a trehalose-synthesis regulator, marks slow-growing cells and contributes to this resistance. I will present these results and discuss them in the context of the evolutionary forces shaping robust and heterogeneous traits. Sayak MukherjeeThe process that warrants the generation of self tolerant peripheral T cells is called the thymocyte selection. During this maturation process, the overtly self reactive as well as unresponsive thymocytes are deleted from the cell population. The thymocytes equipped with T cell receptors (TCRs), capable of responding moderately to the self peptides are allowed to survive. Recently water soluble second messenger, inositol(1,3,4,5) tetrakisphosphate (IP4), has been implicated to play a crucial role in thymocyte positive selection (Huang et al.). It has been suggested that these IP4 molecules regulate the transient activation of the Tec- family protein tyrosine kinase Itk through a competing positive and negative feedbacks. The exact molecular mechanism involved in this feedback is however unclear. It is possible to con- struct more than one model with contrasting molecular mechanisms to explain the present body of experimental observations. This calls for criteria to choose among these models.
Sayak MukherjeeThe process that warrants the generation of self tolerant peripheral T cells is called the thymocyte selection. During this maturation process, the overtly self reactive as well as unresponsive thymocytes are deleted from the cell population. The thymocytes equipped with T cell receptors (TCRs), capable of responding moderately to the self peptides are allowed to survive. Recently water soluble second messenger, inositol(1,3,4,5) tetrakisphosphate (IP4), has been implicated to play a crucial role in thymocyte positive selection (Huang et al.). It has been suggested that these IP4 molecules regulate the transient activation of the Tec- family protein tyrosine kinase Itk through a competing positive and negative feedbacks. The exact molecular mechanism involved in this feedback is however unclear. It is possible to con- struct more than one model with contrasting molecular mechanisms to explain the present body of experimental observations. This calls for criteria to choose among these models. Ilya NemenmanIlya Nemenman, Physics, Emory University
Ilya NemenmanIlya Nemenman, Physics, Emory University Maxim ArtyomovScreens monitoring the effects of deletion, knock-down or over expression of regulatory genes on the expression of their target genes are critical for deciphering the organization of complex regulatory networks. However, since perturbation assays cannot distinguish direct from indirect effects, the derived networks are significantly more complex than the true underlying one. Discovery of the true network organization is a long-standing challenge and several approaches have been developed to infer regulatory networks based on gene expression data. Recent studies indicate [ref] that information obtained from perturbation screens is critical for the ability to identify network structure accurately. This information is typically incorporated into network inference algorithms in two ways: first, the strength of the perturbation effect is translated into interaction confidence values and, second, topological analysis of the experimental networks is performed to find interactions that are most important to preserve network structure and, hence, are more likely to be biological. This underscores importance of having accurate methodology for accurate inference of network topology. In this work we present Exigo, an approach for systematic analysis of network topology. Exigo provides the means to identify core network structure for an input network of any topology with an arbitrary number of activating and inhibiting interactions. We further show that Exigo allows for significant improvement in the network inference. To illustrate this, we constructed a chimeric network inference method that incorporates Exigo into existing inference pipeline [ref], benchmarked it against DREAM challenge networks and found significant improvement in network inference compared to DREAM top performers.
Maxim ArtyomovScreens monitoring the effects of deletion, knock-down or over expression of regulatory genes on the expression of their target genes are critical for deciphering the organization of complex regulatory networks. However, since perturbation assays cannot distinguish direct from indirect effects, the derived networks are significantly more complex than the true underlying one. Discovery of the true network organization is a long-standing challenge and several approaches have been developed to infer regulatory networks based on gene expression data. Recent studies indicate [ref] that information obtained from perturbation screens is critical for the ability to identify network structure accurately. This information is typically incorporated into network inference algorithms in two ways: first, the strength of the perturbation effect is translated into interaction confidence values and, second, topological analysis of the experimental networks is performed to find interactions that are most important to preserve network structure and, hence, are more likely to be biological. This underscores importance of having accurate methodology for accurate inference of network topology. In this work we present Exigo, an approach for systematic analysis of network topology. Exigo provides the means to identify core network structure for an input network of any topology with an arbitrary number of activating and inhibiting interactions. We further show that Exigo allows for significant improvement in the network inference. To illustrate this, we constructed a chimeric network inference method that incorporates Exigo into existing inference pipeline [ref], benchmarked it against DREAM challenge networks and found significant improvement in network inference compared to DREAM top performers. Joanna MaselMaking genes into gene products is subject to predictable errors, each with a phenotypic effect that depends on a normally cryptic sequence. The distribution of fitness effects of these cryptic sequences, like that of new mutations, is bimodal. For example, a cryptic sequence might be strongly deleterious if it causes protein misfolding, or it might have only a minor effect if it preserves the protein fold and tweaks function. Few sequences have effect sizes that fall in between.
Joanna MaselMaking genes into gene products is subject to predictable errors, each with a phenotypic effect that depends on a normally cryptic sequence. The distribution of fitness effects of these cryptic sequences, like that of new mutations, is bimodal. For example, a cryptic sequence might be strongly deleterious if it causes protein misfolding, or it might have only a minor effect if it preserves the protein fold and tweaks function. Few sequences have effect sizes that fall in between. Jon WilkinsWhen natural selection is acting at the level of the the individual phenotype, we expect selection to favor more robust phenotypes. However, selection acting at the level of the gene can undermine adaptation of the individual organism, and lead to the fixation of suboptimal traits. Genomic imprinting, the phenomenon where the pattern of expression of an allele depends on its parental origin, is thought to result from and evolutionary intragenomic conflict, where maternally and paternally inherited alleles favor different optimal phenotypes. Using a simple model, I will illustrate how this can lead to the systematic unravelling of phenotypic robustness.
Jon WilkinsWhen natural selection is acting at the level of the the individual phenotype, we expect selection to favor more robust phenotypes. However, selection acting at the level of the gene can undermine adaptation of the individual organism, and lead to the fixation of suboptimal traits. Genomic imprinting, the phenomenon where the pattern of expression of an allele depends on its parental origin, is thought to result from and evolutionary intragenomic conflict, where maternally and paternally inherited alleles favor different optimal phenotypes. Using a simple model, I will illustrate how this can lead to the systematic unravelling of phenotypic robustness. Emilie Snell-RoodNot available.
Emilie Snell-RoodNot available. David HollowayFruit flies are models for understanding the genetic regulation involved in specifying the complex body plans of higher animals. The head-to-tail (anterior-posterior) axis of the fly (Drosophila) is established in the first hours of development. Maternally supplied factors form concentration gradients which direct embryonic (zygotic) genes where to be activated to express proteins. These protein patterns specify the positions and cell types of the body's tissues. Recent research has shown, comparing between embryos, that the zygotic gene products are much more precisely positioned than the maternal gradients, indicating an embryonic error reduction mechanism. Within embryos, there is the additional aspect that DNA and mRNA operate at very low copy number, and the associated high relative noise has the potential to strongly affect protein expression patterns. In recent work, we have focused on the noise aspects of positional specification within individual embryos, and what molecular mechanisms confer robustness to the process.
David HollowayFruit flies are models for understanding the genetic regulation involved in specifying the complex body plans of higher animals. The head-to-tail (anterior-posterior) axis of the fly (Drosophila) is established in the first hours of development. Maternally supplied factors form concentration gradients which direct embryonic (zygotic) genes where to be activated to express proteins. These protein patterns specify the positions and cell types of the body's tissues. Recent research has shown, comparing between embryos, that the zygotic gene products are much more precisely positioned than the maternal gradients, indicating an embryonic error reduction mechanism. Within embryos, there is the additional aspect that DNA and mRNA operate at very low copy number, and the associated high relative noise has the potential to strongly affect protein expression patterns. In recent work, we have focused on the noise aspects of positional specification within individual embryos, and what molecular mechanisms confer robustness to the process. Ricardo AzevedoEvolution is the movement of populations through a space of genotypes. This space can be modeled as an undirected network connecting genotypes that can be reached through mutation. In this view, the mutational robustness of a genotype is the proportion of its mutational neighbors that are viable. Robustness can facilitate the exploration of genotype networks, or evolvability. Sexual reproduction is also widely believed to promote evolvability, for two reasons. First, because it allows long jumps through genotype space. Second, because it selects for mutational robustness. Here, I show that, depending on the structure of the genotype network, sexual reproduction may not select for the highest mutational robustness, and can actually reduce evolvability.
Ricardo AzevedoEvolution is the movement of populations through a space of genotypes. This space can be modeled as an undirected network connecting genotypes that can be reached through mutation. In this view, the mutational robustness of a genotype is the proportion of its mutational neighbors that are viable. Robustness can facilitate the exploration of genotype networks, or evolvability. Sexual reproduction is also widely believed to promote evolvability, for two reasons. First, because it allows long jumps through genotype space. Second, because it selects for mutational robustness. Here, I show that, depending on the structure of the genotype network, sexual reproduction may not select for the highest mutational robustness, and can actually reduce evolvability.