MBI Videos
Workshop 6: Algebraic Methods in Systems and Evolutionary Biology
-
 Anne ShiuChemical reaction networks taken with mass-action kinetics are dynamical systems governed by polynomial differential equations that arise in systems biology. In general, establishing the existence of (multiple) steady states is challenging, as it requires the solution of a large system of polynomials with unknown coefficients. If, however, the steady state ideal of the system is a binomial ideal, then we show that these questions can be answered easily. This talk focuses on systems with this property, and we say such systems have toric steady states. Our main result gives sufficient conditions for a chemical reaction system to admit toric steady states. Furthermore, we analyze the capacity of such a system to exhibit multiple steady states. An important application concerns the biochemical reaction networks networks that describe the multisite phosphorylation of a protein by a kinase/phosphatase pair in a sequential and distributive mechanism. No prior knowledge of chemical reaction network theory or binomial ideals will be assumed.
Anne ShiuChemical reaction networks taken with mass-action kinetics are dynamical systems governed by polynomial differential equations that arise in systems biology. In general, establishing the existence of (multiple) steady states is challenging, as it requires the solution of a large system of polynomials with unknown coefficients. If, however, the steady state ideal of the system is a binomial ideal, then we show that these questions can be answered easily. This talk focuses on systems with this property, and we say such systems have toric steady states. Our main result gives sufficient conditions for a chemical reaction system to admit toric steady states. Furthermore, we analyze the capacity of such a system to exhibit multiple steady states. An important application concerns the biochemical reaction networks networks that describe the multisite phosphorylation of a protein by a kinase/phosphatase pair in a sequential and distributive mechanism. No prior knowledge of chemical reaction network theory or binomial ideals will be assumed.
This is joint work with Carsten Conradi, Mercedes Pérez Millán, and Alicia Dickenstein. -
 Reka AlbertOver the past five years my group, in collaboration with wet-bench biologists, developed and validated asynchronous Boolean models of several signal transduction networks. Along the way we have encountered obstacles related to the lack of timing knowledge and the large size of the state space. In this talk I will present three methodologies we developed to overcome these obstacles. First, from a comparative analysis of several asynchronous update methods we concluded that updating a single, randomly selected node at each time instant offers the best combination of information and economy.
Reka AlbertOver the past five years my group, in collaboration with wet-bench biologists, developed and validated asynchronous Boolean models of several signal transduction networks. Along the way we have encountered obstacles related to the lack of timing knowledge and the large size of the state space. In this talk I will present three methodologies we developed to overcome these obstacles. First, from a comparative analysis of several asynchronous update methods we concluded that updating a single, randomly selected node at each time instant offers the best combination of information and economy.
Second, we developed a two-step network reduction method which was able to reduce the number of variables by 90% in two different systems without affecting their dynamic behaviors. Third, we proposed an integration of Boolean rules into graph theoretical analysis and showed that this semi-structural method can identify critical signal mediators on par with dynamic models. -
 Aniruddha DattaCancer encompasses various diseases associated with loss of cell-cycle control, leading to uncontrolled cell proliferation and/or reduced apoptosis. Cancer is usually caused by malfunction(s) in the cellular signaling pathways. Malfunctions occur in different ways and at different locations in a pathway. Consequently, therapy design should first identify the location and type of malfunction and then arrive at a suitable drug combination. We consider the growth factor (GF) signaling pathways, widely studied in the context of cancer. Interactions between different pathway components are modeled using Boolean logic gates. All possible single malfunctions in the resulting circuit are enumerated and responses of the different malfunctioning circuits to a 'test' input are used to group the malfunctions into classes. Effects of different drugs, targeting different parts of the Boolean circuit, are taken into account in deciding drug efficacy, thereby mapping each malfunction to an appropriate set of drugs.
Aniruddha DattaCancer encompasses various diseases associated with loss of cell-cycle control, leading to uncontrolled cell proliferation and/or reduced apoptosis. Cancer is usually caused by malfunction(s) in the cellular signaling pathways. Malfunctions occur in different ways and at different locations in a pathway. Consequently, therapy design should first identify the location and type of malfunction and then arrive at a suitable drug combination. We consider the growth factor (GF) signaling pathways, widely studied in the context of cancer. Interactions between different pathway components are modeled using Boolean logic gates. All possible single malfunctions in the resulting circuit are enumerated and responses of the different malfunctioning circuits to a 'test' input are used to group the malfunctions into classes. Effects of different drugs, targeting different parts of the Boolean circuit, are taken into account in deciding drug efficacy, thereby mapping each malfunction to an appropriate set of drugs. -
 Heather HarringtonHow cells make decisions can be investigated using mathematical models. We describe a procedure to decide whether a model is compatible with steady-state data. This method requires no parameter values-- it is based on techniques from algebraic geometry, linear algebra, and optimization. Cellular decisions also depend on where they occur in the cell (e.g., nucleus or cytoplasm). We also find that cellular information processing can be altered by including spatial organization. Borrowing tools from chemical reaction network theory and dynamical systems, we show that the existence of distinct compartments plays a pivotal role in whether a system is capable of multistationarity.
Heather HarringtonHow cells make decisions can be investigated using mathematical models. We describe a procedure to decide whether a model is compatible with steady-state data. This method requires no parameter values-- it is based on techniques from algebraic geometry, linear algebra, and optimization. Cellular decisions also depend on where they occur in the cell (e.g., nucleus or cytoplasm). We also find that cellular information processing can be altered by including spatial organization. Borrowing tools from chemical reaction network theory and dynamical systems, we show that the existence of distinct compartments plays a pivotal role in whether a system is capable of multistationarity. -
 Monika HeinerThis talk reports on our investigations to explore appropriate modelling and analysis techniques for processes evolving simultaneously over time and space, applied to biological systems. Current challenges for modelling in Systems Biology include those associated with issues of complexity and representing systems with multi-scale attributes. A drawback of current modelling approaches, including Petri nets, is their limitation to relatively small networks. We use Stochastic and Continuous Petri Nets to consider continuous time evolution as Markov process or system of Ordinary Differential Equations, and Coloured Petri Nets to statically encode finite discrete space. Combining both concepts yields Coloured Stochastic and Coloured Continuous Petri nets, which allow for directly executable models as well as computational experiments using standard analysis and simulation techniques over very large networks. We illustrate our approach by a couple of case studies, including gradient formation, multistrain bacterial colonies, and planar cell polarity signalling in Drosophila wing.
Monika HeinerThis talk reports on our investigations to explore appropriate modelling and analysis techniques for processes evolving simultaneously over time and space, applied to biological systems. Current challenges for modelling in Systems Biology include those associated with issues of complexity and representing systems with multi-scale attributes. A drawback of current modelling approaches, including Petri nets, is their limitation to relatively small networks. We use Stochastic and Continuous Petri Nets to consider continuous time evolution as Markov process or system of Ordinary Differential Equations, and Coloured Petri Nets to statically encode finite discrete space. Combining both concepts yields Coloured Stochastic and Coloured Continuous Petri nets, which allow for directly executable models as well as computational experiments using standard analysis and simulation techniques over very large networks. We illustrate our approach by a couple of case studies, including gradient formation, multistrain bacterial colonies, and planar cell polarity signalling in Drosophila wing. -
 Ilya ShmulevichRegulatory networks of biomolecular interactions in cells govern virtually all cellular behaviors and functions. Modern measurement technologies are being used to generateinformation on many types of interactions, involving transcriptional and microRNA regulatory networks, signaling networks, and cytokine networks. Temporal measurements of gene and protein expression levels and chromatin modifications, coupled with data fusion strategies that incorporate computational predictions of regulatory mechanisms on the basis of other types of information, such as nucleic acid sequence, can be used to constructdynamical system models of these networks. The analysis and simulation of such models, in conjunction with experimental validation, sheds light on biological function and paves the way toward rational and systematic control strategies intended to drive a diseased system toward a desired state by means of targeted interventions. At the same time, such systems approaches permit new biological observables that reflect system-level behavior that cannot be understood by studying individual sets of interactions. Cellular decision making, maintenance of homeostasis and robustness, sensitivity to diverse types of information in the presence of environmental variability, and coordination ofcomplex macroscopic behavior are examples of such emergent systems-level behavior. Information theoretic approaches combined with elements of dynamical systems theory, such as phase transitions and structure dynamics relationships, are promising frameworks for studying fundamental principles governing living systems at all scales of organization.
Ilya ShmulevichRegulatory networks of biomolecular interactions in cells govern virtually all cellular behaviors and functions. Modern measurement technologies are being used to generateinformation on many types of interactions, involving transcriptional and microRNA regulatory networks, signaling networks, and cytokine networks. Temporal measurements of gene and protein expression levels and chromatin modifications, coupled with data fusion strategies that incorporate computational predictions of regulatory mechanisms on the basis of other types of information, such as nucleic acid sequence, can be used to constructdynamical system models of these networks. The analysis and simulation of such models, in conjunction with experimental validation, sheds light on biological function and paves the way toward rational and systematic control strategies intended to drive a diseased system toward a desired state by means of targeted interventions. At the same time, such systems approaches permit new biological observables that reflect system-level behavior that cannot be understood by studying individual sets of interactions. Cellular decision making, maintenance of homeostasis and robustness, sensitivity to diverse types of information in the presence of environmental variability, and coordination ofcomplex macroscopic behavior are examples of such emergent systems-level behavior. Information theoretic approaches combined with elements of dynamical systems theory, such as phase transitions and structure dynamics relationships, are promising frameworks for studying fundamental principles governing living systems at all scales of organization. -
 Aziz Mithani
Aziz MithaniThe availability of genomes of many closely related bacteria with diverse metabolic capabilities offers the possibility of tracing metabolic evolution on a phylogeny relating the genomes to understand the evolutionary processes and constraints that affect the evolution of metabolic networks. Using simple (independent loss/gain of reactions) or complex (incorporating dependencies among reactions) stochastic models of metabolic evolution, it is possible to study how metabolic networks evolve over time. Here, we describe metabolic network evolution as a discrete space continuous time Markov process and introduce a neighbor-dependent model for the evolution of metabolic networks where the rates with which reactions are added or removed depend on the fraction of neighboring reactions present in the network. The model also allows estimation of the strength of the neighborhood effect during the course of evolution. We present Gibbs samplers for sampling networks at the internal node of a phylogeny and for estimating the parameters of evolution over a phylogeny without exploring the whole search space by iteratively sampling from the conditional distributions of the internal networks and parameters. The samplers are used to estimate the parameters of evolution of metabolic networks of bacteria in the genus Pseudomonas and to infer the metabolic networks of the ancestral pseudomonads. The results suggest that pathway maps that are conserved across the Pseudomonas phylogeny have a stronger neighborhood structure than those which have a variable distribution of reactions across the phylogeny, and that some Pseudomonas lineages are going through genome reduction resulting in the loss of a number of reactions from their metabolic networks.
-
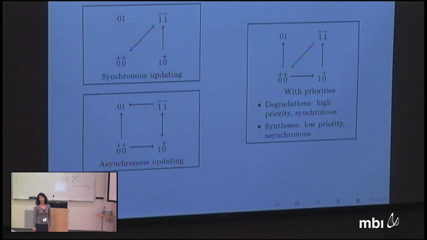 Elisabeth RemyThe logical method proved useful for the modelling of regulatory networks and the analysis of their dynamical properties. It relies on two directed graphs: the regulatory graph, which represents the interactions between regulatory components, each associated with discrete levels of expression (or activity), and the state transition graph, which represents the discrete dynamics of such a model. This discrete modelling framework allows qualitative analyses of the behaviours driven by regulatory networks, based on analytical results or on simulation (i.e. construction of state transition graphs). Although this formalism abstracts and simplifies the biological reality, we still need to cope with challenging issues due to the complexity of ever increasing networks. We present here some results and tools that aim at facilitating the analysis of large networks. In particular, we will show how dynamical properties can be predicted from the presence of particular motifs in the regulatory graph, namely regulatory circuits and combination of such circuits. We will also discuss a method to reduce the model, yet keeping the main features of the dynamics. Finally, we will illustrate these approaches on a generic model of E2F1-dependent apoptosis and cell cycle entries.
Elisabeth RemyThe logical method proved useful for the modelling of regulatory networks and the analysis of their dynamical properties. It relies on two directed graphs: the regulatory graph, which represents the interactions between regulatory components, each associated with discrete levels of expression (or activity), and the state transition graph, which represents the discrete dynamics of such a model. This discrete modelling framework allows qualitative analyses of the behaviours driven by regulatory networks, based on analytical results or on simulation (i.e. construction of state transition graphs). Although this formalism abstracts and simplifies the biological reality, we still need to cope with challenging issues due to the complexity of ever increasing networks. We present here some results and tools that aim at facilitating the analysis of large networks. In particular, we will show how dynamical properties can be predicted from the presence of particular motifs in the regulatory graph, namely regulatory circuits and combination of such circuits. We will also discuss a method to reduce the model, yet keeping the main features of the dynamics. Finally, we will illustrate these approaches on a generic model of E2F1-dependent apoptosis and cell cycle entries.
