MBI Videos
CTW: Mathematical Challenges in Biomolecular/Biomedical Imaging and Visualization
-
 Michael Knopp
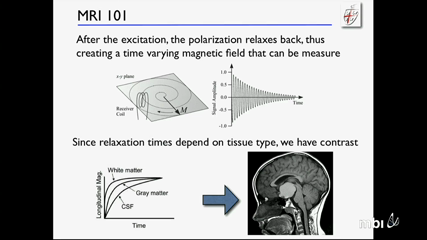
Michael KnoppMichael Knopp's lecture on Imaging Technologies.
-
 Edward Walsh
Edward WalshIn applications such as functional Magnetic Resonance Imaging (fMRI), full, uniformly-sampled Cartesian Fourier (frequency space) measurements are acquired. In order to reduce scan time and increase temporal resolution for fMRI studies, one would like to accurately reconstruct these images from a highly reduced set of Fourier measurements. Compressed Sensing (CS) has given rise to techniques that can provide exact and stable recovery of sparse images from a relatively small set of Fourier measurements. For example, if the images are sparse with respect to their gradient, total-variation minimization techniques can be used to recover those images from a highly incomplete set of Fourier measurements. In this discussion, we propose a new algorithm to further reduce the number of Fourier measurements required for exact or stable recovery by utilizing prior edge information from a high resolution reference image. This reference image (routinely acquired during fMRI studies for anatomic landmarking of activations), or more precisely, the fully sampled Fourier measurements of this reference image, can also be used to provide approximate edge information. By combining this edge information with CS techniques for sparse gradient images, numerical experiments show that we can further reduce the number of Fourier measurements required for exact or stable recovery by a factor of 1.6 3, compared with CS techniques alone, without edge information.
-
 Haomin Zhou
Haomin ZhouWe present a new approach to solve the inverse source problem arising in Fluorescence Tomography (FT). In general, the solution is non-unique and the problem is severely ill-posed. It poses tremendous challenges in image reconstructions. In practice, the most widely used methods are based on Tikhonov-type regularizations, which minimize a cost function consisting of a regularization term and a data fitting term. We propose an alternative method, which overcomes the major difficulties, namely the non-uniqueness of the solution and noisy data fitting, in two separated steps. First we find a particular solution called orthogonal solution that satisfies the data fitting term. Then we add to it a correction function in the kernel space so that the final solution fulfills the regularization and other physical requirements, such as positivity. Moreover, there is no parameter needed to balance the data fitting and regularization terms. In addition, we use an efficient basis to represent the source function to form a hybrid strategy using spectral methods and finite element methods in the algorithm. The resulting algorithm can drastically improve the computation speed over the existing methods. And it is also robust against noise and can greatly improve the image resolutions. This is a joint work with Shui-Nee Chow (Math), Ke Yin (Math) and Ali Behrooz (ECE) from Georgia Tech.
-
 Guohui Song
Guohui SongThe technique of covolutional gridding (CG) has been widely used in applications with non-uniform (Fourier) data such as magnetic resonance imaging (MRI). On the other hand, its error analysis is not fully understood. We consider it as a Fourier frame approximation and present an error analysis accordingly. Moreover, we propose a generalized convolutional gridding (GCG) method as an improved frame approximation.
-
 Larry Carin
Larry CarinLarry Carin's lecture onDictionary Learning and Compressive Imaging.
-
 Adityavikram Viswanathan
Adityavikram ViswanathanFourier reconstruction of piecewise-smooth functions suffers from the familiar Gibbs phenomenon. In Fourier imaging modalities such as magnetic resonance imaging (MRI), this manifests as a loss of detail in the neighborhood of tissue boundaries (due to the Gibbs ringing artifact) as well as long scan times necessitated by the collection of a large number of spectral coefficients (due to the poor convergence properties of the reconstruction method). We present a framework for incorporating edge information - for example, the locations and values of jump discontinuities of a function - in the reconstruction. We show that a simple relationship exists between global Fourier data and local edge information. Knowledge of such edges, either a priori or estimated, enables the synthesis of high-mode spectral coefficients beyond those collected by the MR scanner. Incorporating these synthesized coefficients in an augmented Fourier partial sum reconstruction allows for the generation of scans with significantly improved effective resolution. Further use of spectral re-projection schemes can result in the elimination of all Gibbs artifacts. Numerical results showing accelerated convergence and improved reconstruction quality will be presented.
-
 Niels Volkmann
Niels VolkmannElectron tomography is the most widely applicable method for obtaining 3D information by electron microscopy. It has been realized that electron tomography is capable of providing a complete, molecular resolution three-dimensional mapping of entire proteomes including their detailed interactions. However, to realize this goal, information needs to be extracted efficiently form these tomograms. Owing to low SNR, this task is currently mostly carried out manually. Apart from the subjectivity of the process, its time consuming nature precludes the prospects of high throughput processing. Standard template matching approaches rely on "matched filtering", which can be shown to be a Bayesian classifier as long as the template and the target are nearly identical and the noise is independent and identically distributed, Gaussian, and additive. These conditions are not very well met for electron tomographic reconstructions because the noise is spatially correlated by the reconstruction process and the point spread function. As a consequence, many false hits are generated by this method in areas of high density such as membranes or dense vesicles.
In order to address this challenge, we are developing an alternative method for feature recognition in electron tomography, which is based on the use of reduced representation templates. Reduced representations approximate the target by a small number of anchor points. These anchor points are then used to calculate the scoring function within the search volume. This strategy makes the approach robust against noise and against local variations such as those expected from uneven staining. We recently completed a number of proof-of-concept application of this algorithm for detecting ribosomes in electron tomograms of high-pressure-frozen plastic embedded as well as cryo-sectioned mammalian cell sections. The percentage of false hits for this data drops dramatically from ~50% to under 20% if compared to the matched filter approach. Here, I will describe the principles underlying our approach and will present results obtained from its application.
-
 Shan Zhao
Shan ZhaoRecently, we have introduced a pseudo-time coupled nonlinear partial differential equation (PDE) model for biomolecular solvation analysis. Based on a free energy optimization, a boundary value system is derived to couple a nonlinear Poisson-Boltzmann (NPB) equation for electrostatic potential with a generalized Laplace-Beltrami (GLB) equation defining the biomolecular surface. By introducing a pseudo-time in both processes, a more efficient coupling is achieved through the steady state solution of two nonlinear parabolic PDEs. For the GLB equation, the pseudo-transient continuation is attained via a potential driven geometric flow PDE, which defines a smooth biomolecular surface to characterize the dielectric boundary between biomolecules and the surrounding aqueous environment. The resulting smooth dielectric profile, however, introduce some instability issue in solving the time-dependent NPB equation. This motivates us to develop an operator splitting alternating direction implicit (ADI) scheme, in which the nonlinear instability is completely avoided through analytical integration. To speed up the computation of molecular surface, a new fully implicit ADI scheme is developed too for solving the geometric flow equation. Unconditional stability can be realized by both ADI schemes in solving unsteady NPB and GLB equations separately. In solving a coupled system for real biomolecules and chemical compounds, the proposed numerical schemes are found to be conditionally stable. Nevertheless, the time stability can be maintained by using very large time increments, so that the present biomolecular simulation becomes much faster.
-
 Yang Wang
Yang WangThe classical phase retrieval problem concerns the reconstruction of a function from the magnitude of its Fourier transform. Phase retrieval is an important problem in many applications such as molecular imaging and signal processing. Today the phase retrieval problem has been extended to include more general reconstructions for magnitudes of samples. In this talk I will present an overview of this field, including some of the latest advances both in the theoretical and computational aspects of phase retrieval.
-
 Eric Yuanchang Sun
Eric Yuanchang SunNuclear magnetic resonance spectroscopy (NMR) is heavily employed by chemists and biochemists to study the structures and properties of chemical compounds. The measured data however often contain mixtures of chemicals, subject to changing background and environmental noise. A mathematical problem is to unmix or decompose the measured data into a set of pure or source spectra without knowing the mixing process, a so called blind source separation problem.
In the talk, the speaker shall present algorithms for blind separation of spectral mixtures in noisy conditions. The approach combines geometrical and statistical analysis of the data, the geometric approach is based on the vertexes and facets identification of cone structures of the data, while the statistical approach is on decomposing fitting errors when partial knowledge of the source spectra is available. Computational results on data from NMR, Raman spectroscopy and differential optical absorption spectroscopy show the applicability of the methods. This is joint work with Jack Xin from UC Irvine.
-
 Arthur Olson
Arthur OlsonBiology has become accessible to an understanding of processes that span from atom to organism. As such we now have the opportunity to build a spatio-temporal picture of living systems at the molecular level. In our recent work we attempt to create, interact with, and communicate physical representations of complex molecular environments. I will discuss the challenges and demonstrate three levels of interaction with protein interactions: 1) human perceptual and cognitive interaction with complex structural information; 2) interaction and integration of multiple data sources to construct cellular environments at the molecular level; and 3) interaction of software tools that can bridge the disparate disciplines needed to explore, analyze and communicate a holistic molecular view of living systems.
In order to increase our understanding and interaction with complex molecular structural information we have combined two evolving computer technologies, solid printing and augmented reality. We create custom tangible molecular models and track their manipulation with real-time video, superimposing text and graphics onto the models to enhance their information content and to drive interactive computation. We have recently developed automated technologies to construct the crowded molecular environment of living cells from structural information at multiple scales as well as bioinformatics information on levels of protein expression and other data. We can populate cytoplasm, membranes, and organelles within the same structural volume to generate cellular environments that synthesize our current knowledge of such systems.
The communication of complex structural information requires extensive scientific knowledge as well as expertise in creating clear visualizations. We have developed a method of combining scientific modeling environments with professional grade 3D modeling and animation programs such as Maya, Cinema4D and Blender. This gives both scientists and professional illustrators access to the best tools to communicate the science and the art of the molecular cell.
Gillet, A., Sanner, M., Stoffler, D., Olson, A.J. (2005) Tangible interfaces for structural molecular biology. Structure:13:483-491.
Johnson, G.T., Autin, L., Goodsell, D.S., Sanner, M.F., Olson A.J. (2011) ePMV Embeds Molecular Modeling into Professional Animation Software Environments. Structure 19(3):293-303.
-
 Yiying Tong
Yiying TongWe present a method for computing "choking" loops - a set of surface loops that describe the narrowing of the volumes inside/outside of the surface and extend the notion of surface homology and homotopy loops. The intuition behind their definition is that a choking loop represents the region where an offset of the original surface would get pinched. Our generalized loops naturally include the usual $2g$ handles/tunnels computed based on the topology of the genus-$g$ surface, but also include loops that identify chokepoints or bottlenecks, i.e., boundaries of small membranes separating the inside or outside volume of the surface into disconnected regions. Based on persistent homology theory, our definition builds on a measure to topological structures, thus providing resilience to noise and a well-defined way to determine topological feature size.
