MBI Videos
Workshop 4: Tumor Heterogeneity and the Microenvironment
-
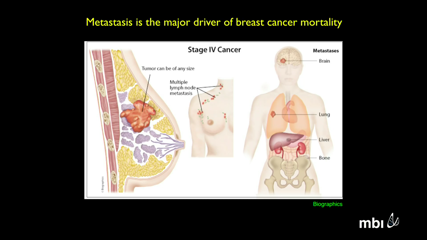 Andrew Ewald
Andrew EwaldAbstract not submitted.
-
 Michael Ostrowski
Michael OstrowskiAbstract not submitted.
-
 Trevor Graham
Trevor GrahamCarcinogenesis is an evolutionary process; establishing the prognosis for a cancer therefore requires predicting the future course of cancer evolution. The same is true in pre-cancerous conditions: the risk of developing cancer is determined by how the pre-cancerous lesion is evolving.
The level of heterogeneity within a population measures the evolvability of the population: if there is no diversity natural selection cannot operate, whereas diverse populations are likely to contain well-adapted individuals that can prosper in changing environments. Consequently, quantification of within-tumour heterogeneity is likely to be a proxy-measure of the rate of the underlying evolutionary process that drives carcinogenesis, and so be an effective prognostic marker. In this talk, I will describe how we have measured within-tumour diversity, both genetically and phenotypically, to successfully determine prognosis in both established cancers and in premalignant lesions.
In addition, I will describe how we begun to search for the most prognostic measures of intra-tumour heterogeneity by constructing simple computational models of cancer development, and using the models to perform an exhaustive search of possible heterogeneity measures.
-
 Simon Hayward
Simon HaywardBoth stromal and epithelial cells in tumors exhibit marked heterogeneity that can affect communication within and between tissue layers resulting in alterations in cell proliferation, survival and death. Many of these interactions are cooperative rather than competitive. The various signaling axes represent a complex network of altered ligand/receptor availability that can be modified to alter growth and progression. A more complete understanding of cellular complexity in both stromal and epithelial tissues should allow us to model these intercellular communications with a view to identifying key nodes that can be coordinately modulated to restrict tumor growth.
-
 Mary Helen Barcellos-Hoff
Mary Helen Barcellos-HoffBoth clinical and experimental data show that the stroma is highly involved in early malignancy, supporting the idea of reciprocal evolution of the malignant cell and the tumor microenvironment. Although it is clear that stroma composition and signaling is altered in cancer, less is known about how and when stroma contributes to carcinogenesis and how carcinogens, like radiation, might alter these processes. To investigate how tumor diversity evolves in the context of different physiological states (e.g. as a function of age and ionizing radiation exposure) we developed a mammary chimera model in which histologically and genomically diverse cancer originate from a p53 null epithelium orthotopically transplanted to a syngeneic wildtype host. We use expression profiling of tissue and tumors to identify the biological ‘bookends’, those processes initiated in normal tissue that appear to contribute to the development and are recapitulated in particular types of tumors. These studies show that the spectrum of mammary tumor types is strongly influenced by the host biology, opening new perspective on the drivers of aggressive cancer. The mounting evidence from these and other studies that cancer results from a systemic failure in which cells other than those with oncogenic alterations determine the frequency and type of clinical cancer is changing the cancer paradigm. To account for this, we propose that the tumor microenvironment is built through rate-limiting steps during multi-stage carcinogenesis [Barcellos-Hoff, 2013 #18417]. In this model, construction of a ‘pre-cancer niche’ is a necessary early step required for initiated cells to survive and evolve; subsequent niche expansion and maturation accompany promotion and progression respectively. The model postulates that cancer cell survival and proliferation is as much a function of the successful niche construction as it is of the natural selection for specific cancer cell mutations. Consequently, cancer represents an emergent property that requires a comprehensive analysis of the cell-cell interactions during the course of carcinogenesis. Moreover, in contrast to initiation, which is stochastic by nature, niche construction represents a robust target for native immunosuppression and a potent target for cancer prevention.
-
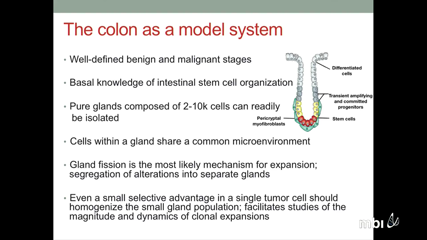 Christina Curtis
Christina Curtis
What happens in the early and still undetectable human malignancy is unknown because direct observations are impractical. Here I will describe a novel “Big Bang� model, whereby a tumor grows predominantly as a single expansion producing numerous intermixed sub-clones, which are not subject to stringent clonal selection. In this model, both public and most detectable private mutations arise during the earliest phase of tumor growth. Multi-scale genomic profiling of 349 individual glands sampled from 15 colorectal tumors revealed the absence of selective sweeps, uniformly high intra-tumor heterogeneity, and sub-clone mixing in distant tumor regions, as postulated by the Big Bang. By integrating the data in a spatial model of tumor growth and statistical inference framework we also verified the most striking prediction of our model, namely that most detectable intra-tumor heterogeneity originates from private alterations acquired early during growth, and not from the later expansion of selected sub-clones. Hence, early sub-clones define the genomic profile of colorectal carcinomas and advanced adenomas, whereas potentially dangerous late-arising sub-clones will go undetected. Moreover, our results suggest that sub-clone mixing may be a biomarker of malignant potential. This new model provides a quantitative framework that explains the origins of intra-tumor heterogeneity and tumor growth dynamics with significant clinical implications for treatment resistance and metastatic progression, as I will discuss.
-
 Joseph Costello
Joseph CostelloTumor recurrence is a leading cause of cancer mortality. Therapies for recurrent disease may fail, at least in part, because the genomic alterations driving the growth of recurrences are distinct from those in the initial tumor. At diagnosis, low grade brain tumors have similar histology and driver mutations, but following surgical resection their clinical behavior is highly variable. Infiltrating tumor cells comprising the residual disease may remain indolent for more than a decade after the surgical resection, or may rapidly transform into an aggressive rapidly growing malignant tumor. Adjuvant chemotherapies such as temozolomide (TMZ) are frequently used, especially in cases with subtotal surgical resections or when deferring the use of radiation therapy is preferable. We used genome and epigenome sequencing technologies to infer patterns of intratumoral heterogeneity and tumor evolution over time. In this presentation I will discuss new insight into diverging genetic pathways arising from initially similar tumors, and the profound influence of chemotherapy in shaping tumor aggressiveness.
-
 Robert Gatenby
Robert GatenbyGenetic and epigenetic changes in cancer cells are typically divided into "drivers" and "passengers" Drug development strategies target driver mutations, but inter- and intra-tumoral heterogeneity usually results in emergence of resistance. Here we model intratumoral evolution in the context of a fecundity/survivorship trade-off. Simulations demonstrate the fitness value, of any genetic change is not fixed but dependent on evolutionary triage governed by initial cell properties, current selection forces, and prior genotypic/phenotypic trajectories. We demonstrate spatial variations in molecular properties of tumor cells are the result of changes in environmental selection forces such as blood flow. Simulated therapies targeting fitness-increasing (driver) mutations usually decrease the tumor burden but almost inevitably fail due to population heterogeneity. An alternative strategy targets gene mutations that are never observed. Because up or down regulation of these genes unconditionally reduces cellular fitness, they are eliminated by evolutionary triage but can be exploited for targeted therapy.
-
 Daniel Worthley
Daniel WorthleyThe tumor microenvironment presents an exciting opportunity for innovative therapeutic approaches to cancer. The origin and exact biological contribution of the peritumoral connective tissue at the primary and metastatic sites, however, is still uncertain. Just as one would not assess the average of “hematopoietic� contribution to the tumor microenvironment without considering distinct cell types, so too lumping together all “peritumoral mesenchyme� or “cancer-associated fibroblasts� on the basis of broad markers is likely to oversimplify a diverse connective-tissue contribution to cancer. In this presentation I will discuss the types of connective tissue cells in cancer. I will outline recent studies addressing the biological function of these cells in cancer and discuss our own research on the heterogeneity of connective tissue stem cells in the intestine, the bone and in several cancer models. Understanding the biological heterogeneity of mesenchymal cells in cancer will provide new opportunities for targeted cancer prevention and therapy.
-
 Alexander Anderson
Alexander AndersonHeterogeneity in cancer is an observed fact, both genetically and phenotypically. Intercellular variation is seen at all scales and stages of development, and has significant implications for prognosis. At present, our understanding of this heterogeneity is mainly restricted to the genetic scale with little information regarding the relationship between genetic and phenotypic heterogeneity. Further, little is known about how cells alter their microenvironment or how these changes drive selection and feedback to further drive cancer evolution.
Strong selective pressures imposed by a milieu of microenvironmental factors combined with high profileration rates and high mutation rates inevitably lead to the rapid emergence of resistance to therapy. Hence, the failure of cancer therapies is often attributed to Darwinian evolution. To understand and predict cancer evolution we must understand not only the mutations which drive evolution but also the mechanisms through which these mutations manifest themselves in phenotypic change. Thus, our success in predicting cancer progression and designing effective therapy is contingent on understanding the junction at which genes and environment meet to produce phenotypes, the genotype-phenotype (GP) map.
Experimental studies have revealed the complexity inherent within the GP-map which is responsible for the difficulty in predicting evolution; many genotypes produce identical phenotypes and further many phenotypes can emerge from a single phenotype. Indeed, recent experimental evidence shows that this mapping produces phenotypic heterogeneity through a variety of genetic and non--genetic mechanisms. Heterogeneity can be driven through phenotypic plasticity, the phenomenon whereby isogenic cells in different environments display different phenotypes. Further, isogenic cells in identical environments can display phenotypic heterogeneity which is the manifestation of intra--cellular noise amplified through the complex machinery of the cell signalling pathways. In this talk I will present a collection of related mathematical models which explore genetic, environmental, phenotypic and morphological heterogeneity through the unifying lens of the GP-map, outline the key mechanisms which could be responsible for generating adaptive phenotypes -- mutation, plasticity and stochasticity -- and explore the implications of these mechanisms for developing novel and effective cancer therapies.
-
 Philip Gerlee
Philip GerleeCancer cells are known to modify their micro-environment such that it can sustain a larger population, or, in ecological terms, they construct a niche which increases the carrying capacity of the population. It hashowever been argued that niche construction, which benefits all cells in the tumour, would be selected against since cheaters could reap the benefits without paying the cost. We have investigated the impact of niche specificity on tumour evolution using an individual based model of breast tumour growth, in which the carrying capacity of each cell consists of two components: an intrinsic, subclone-specific part and a contribution from all neighbouring cells. Analysis of the model shows that the ability of a mutant to invade a resident population depends strongly on the specificity. When specificity is low selection is mostly on growth rate, while high specificity shifts selection towards increased carrying capacity. Further, we show that the long-term evolution of the system can be predicted using adaptive dynamics. By comparing the results from a spatially structured vs. well-mixed population we show that spatial structure restores selection for carrying capacity even at zero specificity, which poses a possible solution to the niche construction dilemma. Lastly, we show that an expanding population exhibits spatially variable selection pressure, where cells at the leading edge exhibit higher growth rate and lower carrying capacity than those at the centre of the tumour.
