MBI Videos
Workshop 7: Stem Cells, Development, and Cancer
-
 Paul MacklinIn diverse fields spanning developmental biology, tissue engineering, and cancer, dynamical interactions between large multicellular populations and the microenvironment shape the emergent behaviors of the larger systems, often in surprising ways. While mathematical models can help to understand these systems, 3-D simulation studies often require solving for the dynamics of 105 or more cells, along with several diffusing growth substrates and signaling factors. Such simulations are challenging, generally requiring either complex parallelization on supercomputers, or restriction to smaller systems of 103 or 104 cells in 2-D or very small 3-D domains. Moreover, largely incompatible data formats have impeded the development of robust tools for 3-D visualization and data analysis, further slowing systematic investigations. We will discuss our work to address these problems with parallelized open source codes designed for desktop computers or single HPC compute nodes. We will introduce PhysiCell: an agent-based model that simulates cell cycling, apoptosis, necrosis, volume changes, and biomechanics-based movement in 3-D systems of 105 to 106 cells. We will introduce BioFVM: a finite volume solver for diffusive transport in large 3-D tissues (5+ diffusing substrates on 106 or more voxels: 5-10 mm3 at 20 �m resolution). We will discuss MultiCellDS: a draft data standard for cell phenotype and multicellular simulation data. And we will discuss our efforts to connect these tools to simulate breast cancer and colon cancer metastases in the liver. It is our hope that this work will help seed an ecosystem of compatible computational tools and experimental data. At the close of the talk, we will open the floor to discussion on how these tools can be adapted to meet the needs of the stem cell modeling community. See mbi2015.MathCancer.org for more information.
Paul MacklinIn diverse fields spanning developmental biology, tissue engineering, and cancer, dynamical interactions between large multicellular populations and the microenvironment shape the emergent behaviors of the larger systems, often in surprising ways. While mathematical models can help to understand these systems, 3-D simulation studies often require solving for the dynamics of 105 or more cells, along with several diffusing growth substrates and signaling factors. Such simulations are challenging, generally requiring either complex parallelization on supercomputers, or restriction to smaller systems of 103 or 104 cells in 2-D or very small 3-D domains. Moreover, largely incompatible data formats have impeded the development of robust tools for 3-D visualization and data analysis, further slowing systematic investigations. We will discuss our work to address these problems with parallelized open source codes designed for desktop computers or single HPC compute nodes. We will introduce PhysiCell: an agent-based model that simulates cell cycling, apoptosis, necrosis, volume changes, and biomechanics-based movement in 3-D systems of 105 to 106 cells. We will introduce BioFVM: a finite volume solver for diffusive transport in large 3-D tissues (5+ diffusing substrates on 106 or more voxels: 5-10 mm3 at 20 �m resolution). We will discuss MultiCellDS: a draft data standard for cell phenotype and multicellular simulation data. And we will discuss our efforts to connect these tools to simulate breast cancer and colon cancer metastases in the liver. It is our hope that this work will help seed an ecosystem of compatible computational tools and experimental data. At the close of the talk, we will open the floor to discussion on how these tools can be adapted to meet the needs of the stem cell modeling community. See mbi2015.MathCancer.org for more information. -
 Arianna Bianchi
Arianna BianchiPrimary tumors infrequently lead to the demise of cancer patients; rather, mortality and a significant degree of morbidity result from the growth of secondary tumors in distant organs (metastasis). Malignant tumors release both lymph- and angio- genic factors, through two specific processes termed lymphangiogenesis and angiogenesis, respectively. In addition, recent experimental evidence shows that tumors initiate their own innervation by the release of neurotrophic factors (neoneurogenesis). The relationship between tumor progression and the nervous system is a complex and poorly understood part of cancer pathogenesis. It is likely that this process is regulated by a multitude of factors in the tumor/nerve microenvironment; these pathways are even further complicated by treatment and disease history as well as other genetic and socioeconomic factors. It is therefore important to study the interactions between the nervous system and tumor cells through mathematical/computational modelling: in this way we will take into account the most significant elements of the plethora of interacting pathways regulating this process. The present work is a first attempt to model the neurobiological aspect of cancer development through a system of differential equations.
NOTE: This is a joint work with Dr Georgios Lolas, TU Dresden (Germany).
-
 Alexandra JilkineRecent evidence suggests that, like many normal tissues, many cancers are maintained by a small population of cancer stem cells that divide indefinitely to produce more differentiated cancerous cells.
Alexandra JilkineRecent evidence suggests that, like many normal tissues, many cancers are maintained by a small population of cancer stem cells that divide indefinitely to produce more differentiated cancerous cells.
Tissues, however, contain many more differentiated cells than stem cells, and mutations may cause such cells to "dedifferentiate" into a stem-like state.
We study the effects of dedifferentiation on the time to cancer onset and found that the effect of dedifferentiation depends critically on how stem cell numbers are controlled by the body. If homeostasis is very tight (due to all divisions being asymmetric), then dedifferentiation has little effect, but if homeostatic control is looser (allowing both symmetric and asymmetric divisions), then dedifferentiation can dramatically hasten cancer onset and lead to exponential growth of the cancer stem cell population. We also consider effects of various negative feedback loops from the progenitor population on regulation of homeostasis.
Our results suggest that dedifferentiation may be a very important factor in cancer and that more study of dedifferentiation and stem cell control is necessary to understand and prevent cancer onset. -
 Heiko EnderlingGlioblastoma multiforme (GBM) is one of the most aggressive human malignancies with a poor patient prognosis. Ionizing radiation either alone or adjuvant after surgery is part of standard treatment for GBM but remains primarily noncurative. The mechanisms underlying tumor radioresistance are manifold and, in part, accredited to a special subpopulation of tumorigenic cells. The so-called glioma stem cells (GSC) are bestowed with the exclusive ability to self-renew and repopulate the tumor and have been reported to be less sensitive to radiation- induced damage through preferential activation of DNA damage checkpoint responses and increased capacity for DNA damage repair. During each fraction of radiation, non–stem cancer cells (CC) die and GSCs become enriched and potentially increase in number, which may lead to accelerated repopulation. We propose a cellular Potts model that simulates the kinetics of GSCs and CCs in glioblastoma growth and radiation response. We parameterize and validate this model with experimental data of the U87-MG human glioblastoma cell line. Simulations are conducted to estimate GSC symmetric and asymmetric division rates and explore potential mechanisms for increased GSC fractions after irradiation. Simulations reveal that in addition to their higher radioresistance, a shift from asymmetric to symmetric division or a fast cycle of GSCs following fractionated radiation treatment is required to yield results that match experimental observations. We hypothesize a constitutive activation of stem cell division kinetics signaling pathways during fractionated treatment, which contributes to the frequently observed accelerated repopulation after therapeutic irradiation.
Heiko EnderlingGlioblastoma multiforme (GBM) is one of the most aggressive human malignancies with a poor patient prognosis. Ionizing radiation either alone or adjuvant after surgery is part of standard treatment for GBM but remains primarily noncurative. The mechanisms underlying tumor radioresistance are manifold and, in part, accredited to a special subpopulation of tumorigenic cells. The so-called glioma stem cells (GSC) are bestowed with the exclusive ability to self-renew and repopulate the tumor and have been reported to be less sensitive to radiation- induced damage through preferential activation of DNA damage checkpoint responses and increased capacity for DNA damage repair. During each fraction of radiation, non–stem cancer cells (CC) die and GSCs become enriched and potentially increase in number, which may lead to accelerated repopulation. We propose a cellular Potts model that simulates the kinetics of GSCs and CCs in glioblastoma growth and radiation response. We parameterize and validate this model with experimental data of the U87-MG human glioblastoma cell line. Simulations are conducted to estimate GSC symmetric and asymmetric division rates and explore potential mechanisms for increased GSC fractions after irradiation. Simulations reveal that in addition to their higher radioresistance, a shift from asymmetric to symmetric division or a fast cycle of GSCs following fractionated radiation treatment is required to yield results that match experimental observations. We hypothesize a constitutive activation of stem cell division kinetics signaling pathways during fractionated treatment, which contributes to the frequently observed accelerated repopulation after therapeutic irradiation. -
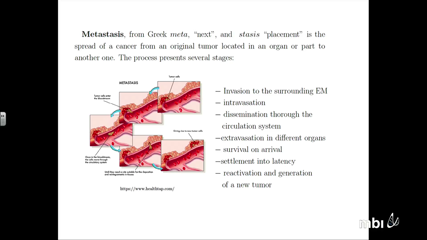 Jose Ignacio Tello
Jose Ignacio Tello -
 Jan PoleszczukCells of different organs at different ages have an intrinsic set of kinetics that dictates their behavior. Transformation into cancer cells will inherit these kinetics that determine initial cell and tumor population progression dynamics. Subject to genetic mutation and epigenetic alterations, cancer cell kinetics can change and favorable alterations that increase cellular fitness will manifest themselves and accelerate tumor progression. We set out to investigate the emerging intratumoral heterogeneity and to determine the evolutionary trajectories of the combination of cell-intrinsic kinetics that yield aggressive tumor growth. We develop a cellular automaton model that tracks the temporal evolution of the malignant subpopulation of so-called cancer stem cell, as these cells are exclusively able to initiate and sustain tumors. We explore orthogonal cell traits including cell migration to facilitate invasion, spontaneous cell death due to genetic drift after accumulation of irreversible deleterious mutations, symmetric cancer stem cell division that increases the cancer stem cell pool, and telomere length and erosion as a mitotic counter for inherited non-stem cancer cell proliferation potential. Our study suggests that cell proliferation potential is the strongest modulator of tumor growth. Early increase in proliferation potential yields larger populations of CC that compete with CSC and thus inhibit CSC division while a reduction in proliferation potential loosens such inhibition and facilitates frequent CSC division. The subpopulation of cancer stem cells in itself becomes highly heterogeneous dictating population level dynamics that vary from long-term dormancy to aggressive progression. Our study suggests that the clonal diversity that is captured in single tumor biopsy samples represents only a small proportion of the total number of phenotypes.
Jan PoleszczukCells of different organs at different ages have an intrinsic set of kinetics that dictates their behavior. Transformation into cancer cells will inherit these kinetics that determine initial cell and tumor population progression dynamics. Subject to genetic mutation and epigenetic alterations, cancer cell kinetics can change and favorable alterations that increase cellular fitness will manifest themselves and accelerate tumor progression. We set out to investigate the emerging intratumoral heterogeneity and to determine the evolutionary trajectories of the combination of cell-intrinsic kinetics that yield aggressive tumor growth. We develop a cellular automaton model that tracks the temporal evolution of the malignant subpopulation of so-called cancer stem cell, as these cells are exclusively able to initiate and sustain tumors. We explore orthogonal cell traits including cell migration to facilitate invasion, spontaneous cell death due to genetic drift after accumulation of irreversible deleterious mutations, symmetric cancer stem cell division that increases the cancer stem cell pool, and telomere length and erosion as a mitotic counter for inherited non-stem cancer cell proliferation potential. Our study suggests that cell proliferation potential is the strongest modulator of tumor growth. Early increase in proliferation potential yields larger populations of CC that compete with CSC and thus inhibit CSC division while a reduction in proliferation potential loosens such inhibition and facilitates frequent CSC division. The subpopulation of cancer stem cells in itself becomes highly heterogeneous dictating population level dynamics that vary from long-term dormancy to aggressive progression. Our study suggests that the clonal diversity that is captured in single tumor biopsy samples represents only a small proportion of the total number of phenotypes.
