-
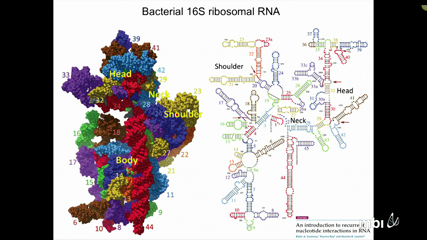
Neocles Leontis
-
Karin Musier-Forsyth
The 5' untranslated region (5'-UTR) is a highly conserved region of retroviral RNA genomes responsible for regulating many steps of the retroviral lifecycle including viral RNA dimerization, packaging, initiation of reverse transcription, transcriptional regulation, and splicing. A complete understanding of the mechanisms controlling retroviral replication requires structural characterization of this RNA. Unfortunately, its large size and conformational flexibility renders common methods of solving structures, such as X-ray crystallography and NMR exceedingly difficult. Here, we use a solution technique, small-angle X-ray scattering (SAXS), coupled with computational molecular modeling and structure probing, to characterize RNAs (100-350 nucleotides in length) derived from the 5'-UTR of HIV-1 (NL4-3 and MAL isolates), RSV, SIV, and HTLV-1. Similarities and differences in their packaging signals, the presence of tRNA structural mimicry, conformational switches upon dimerization and primer annealing, and length-dependent changes in global conformation will all be discussed.
-
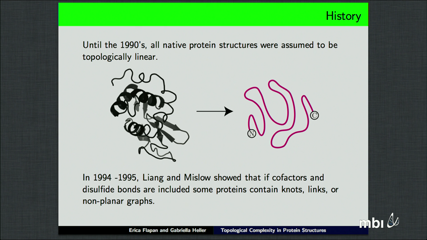
Erica Flapan
For DNA molecules, topological complexity occurs exclusively as the result of knotting or linking of the polynucleotide backbone. By contrast, while a few knots and links have been found within the polypeptide backbones of some protein structures, non-planarity can also result from the connectivity between a polypeptide chain and inter- and intra-chain linking via cofactors and disulfide bonds. In this talk, we survey the known types of knots, links, and non-planar graphs in protein structures with and without including such bonds and cofactors. Then we present new examples of protein structures containing Möbius ladders and other non-planar graphs as a result of these cofactors. Finally, we propose hypothetical structures illustrating specific disulfide connectivities that would result in the key ring link, the Whitehead link and the 5-1 knot, the latter two of which have thus far not been identified within protein structures.
-

Alexander Vologodskii
Type II DNA topoisomerases can change DNA topology by catalyzing the passing of one double-stranded DNA segment through another. In 1997 Rybenkov et al. unexpectedly found that the enzymes can greatly reduce, up to hundred times, the fractions of knotted and linked circular DNA molecules comparing with the corresponding equilibrium values. The phenomenon of topology simplification attracted a lot of attention because it was very difficult to understand how small enzymes could determine topology of large DNA molecules. It seems clear now that the only way for the topoisomerases to achieve topology simplification is to use the fact that the probability of some specific local conformations of DNA segments depends on DNA topology. Although great progress has been made in understanding the phenomenon, some features of it are not explained by the existing models. To eliminate the discrepancy with the experimental data we suggest here a new model of the enzyme action.
-

Konstantin Mischaikow
No abstract has been provided.
-

Mariel Vazquez
Cellular processes such as replication, recombination, and packing change the topology of DNA. Controlling these changes is key to ensuring stability inside the cell. We use topological and computational methods to study the action of enzymes that simplify the topology of DNA during replication. I will review these methods and will expand on some thoughts on DNA folding.
-
Yann Ponty
The prediction of the most stable, prevalent and/or functional structure adopted by an RiboNucleic Acid molecule (RNA) is an old, yet very much ongoing, challenge of computational biology. Currently available computational methods, such as MFold or RNAfold, somehow artificially restrict their search space to tree-like conformations, the secondary structures. However, such a definition intrinsically discards complex topological motifs that are both observed in experimentally-determined structures, essential for the functions performed by the molecule, and conserved throughout the evolution. In this talk, I will review two decades of works aiming at characterizing the complexity of minimizing the free energy of a given RNA molecule, while allowing (limited subsets of) pseudoknots/crossing interactions.
The general hardness of the associated computational problems motivates the development of novel parameterized-complexity approaches and heuristics, as further illustrated by the follow up talk by H Orland.
This is a joint work in collaboration with S. Sheikh (Bloomberg R&D, USA) and R Backofen (Uni. Freiburg, Germany).
-

David Murrugarra
Boolean networks are an important class of computational models for molecular interaction networks. Boolean canalization, a type of hierarchical clustering of the inputs of a Boolean function, has been extensively studied in the context of network modeling where each layer of canalization adds a degree of stability in the dynamics of the network. Recently, dynamic network control approaches have been used for the design of new therapeutic interventions and for other applications such as stem cell reprogramming. This talk will discuss the role of canalization in the control of Boolean molecular networks. A method for identifying the potential control edges in the wiring diagram of a network for avoiding undesirable state transitions will be presented. The method is based on identifying appropriate input-output combinations on undesirable transitions that can be modified using the edges in the wiring diagram of the network. Moreover, a method for estimating the number of changed transitions in the state space of the system as a result of an edge deletion in the wiring diagram will be presented. These two complementary methods can help in the selection of appropriate controllers such as for minimizing the side effects resulting from an edge manipulation.
-

Yusu Wang
I will talk about the use of topological terrain metaphors for (biological) data visualization and analysis. I will in praticular describe two software we developed: Denali, a generic tool for visualizing tree-like structures (such as clustering trees) using topological terrain metaphors, as well as Ayla, a specialized visual analytic tool for exploring molecular simulation data. This is joint work with J. Eldridge, W. Harvey, M. Belkin, T.-P. Bremer, C. Li, I. Park, V. Pascucci and O. Ruebel.
-

Robert Eisenberg
Chemical reactions are described and analyzed using conservation of mass and the law of mass action. The conservation of mass does not imply the conservation of electric current, as can easily be seen by in the reaction A •••• B •••• C where IAB ≠IBC . The two reactions involve different rate constants, that are customarily independent, so the currents cannot be equal under more than one condition! Electric forces are very very much stronger than diffusion forces: one percent change in net charge produces a force large enough to lift the earth; one per cent change in mass has hardly any effect. I argue that chemical models cannot transferable (with one set of parameters) if they do not satisfy conservation of current. I argue that conservation of current must be exact in models of chemical reactions in all conditions, locations, and times because the ‘current’ defined in Maxwell’s equations cannot be stored, at all. My colleagues and I are trying to construct such models, following the lead of colleagues in semiconductor and computational electronics, who have done this for years.
-

Douglas Turner
Less than 5% of the human genome codes for protein, but about 90% codes for transcribed RNA. Structures and functions for much of this RNA are not known. Moreover, it appears to be more difficult to determine and to predict structures of RNAs than structures of proteins. For proteins and RNA, 3D predictions are made by both "knowledge based" and "quantum mechanical based" force fields. NMR experiments on simple RNA systems can provide benchmarks for testing methods to predict 3D structure. Results from NMR experiments and molecular dynamics simulations will be presented for single stranded, unpaired tetramers and for base paired structures of RNA. Comparisons reveal strengths and weaknesses of force fields.
-

Alexander Grosberg
In the talk, I plan to review the current status of the controversial idea that knots in a macromolecule tighten themselves for entropic reasons.
-
Kelin Xia
Proteins are the most important biomolecules for living organisms. The understanding of protein structure, function, dynamics, and transport is one of the most challenging tasks in biological science. We have introduced persistent homology for extracting molecular topological fingerprints (MTFs) based on the persistence of molecular topological invariants. MTFs are utilized for protein characterization, identification, and classification. Both all-atom and coarse-grained representations of MTFs are constructed. On the basis of the correlation between protein compactness, rigidity, and connectivity, we propose an accumulated bar length generated from persistent topological invariants for the quantitative modeling of protein flexibility. To this end, a correlation matrix-based filtration is developed. This approach gives rise to an accurate prediction of the optimal characteristic distance used in protein B-factor analysis. Finally, MTFs are employed to characterize protein topological evolution during protein folding and quantitatively predict the protein folding stability. An excellent consistence between our persistent homology prediction and molecular dynamics simulation is found. This work reveals the topology-function relationship of proteins.
-

Miranda Holmes-Cerfon
Particles in soft-matter systems (such as colloids) tend to have very short-range interactions, so traditional theories, that assume the energy landscape is smooth enough, will struggle to capture their dynamics. We propose a new framework to look at such particles, based on taking the limit as the range of the interaction goes to zero. In this limit, the energy landscape is a set of geometrical manifolds plus a single control parameter, while the dynamics on top of the manifolds are given by a hierarchy of Fokker-Planck equations coupled by "sticky" boundary conditions. We show how to compute dynamical quantities such as transition rates between clusters of hard spheres, and then show this agrees quantitatively with experiments on colloids. We hope this framework is useful for modelling other systems with geometrical constraints, such as those that arise in biology.
-

Alan Rein
Recombinant HIV-1 Gag protein, purified from bacteria, is a soluble protein, but assembles into virus-like particles (VLPs) in a defined in vitro system upon addition of nucleic acid (NA). We have tried to understand how NA-binding promotes assembly. Experiments with Gag-leucine zipper chimeric proteins suggest that when Gag attains a high local concentration, it undergoes a change which primes it for assembly; cooperative NA-binding would be one way it could reach a high local concentration. We have focused on the SP1 region of Gag, which lies between the CA domain (principally responsible for Gag-Gag interactions in VLPs) and the NC domain (principally responsible for NA-binding). Many subtle mutations in SP1 cause drastic disruption of VLP assembly. If the SP1 sequence were folded into an α-helix, the helix would be amphipathic, with a polar face and a hydrophobic face. We have found that a peptide representing SP1 folds into a helix when it is at high concentration; presumably, at these concentrations, helices can aggregate and bury their hydrophobic faces within helix bundles.
We now report that small proteins consisting of SP1 fused to a dimerizing leucine zipper form discrete tetramers in solution. However, many mutations in these SP-zipper chimeras cause them to form dimers rather than tetramers; therefore, SP1-SP1 interactions are responsible for the association of the zipper-induced dimers into tetramers. As these specific mutations also disrupt assembly by Gag, the same SP1-SP1 interactions are also significant in VLP assembly. We hypothesize that folding of SP1 into a helix primes Gag for VLP assembly, perhaps by exposing new interfaces in the CA domain. Information on the structure of these SP1 bundles will be presented.
-

Peijun Zhang
My research program is interested in understanding the structural mechanisms of macromlecular assemblies using an integrated approach by combining three-dimensional cryo-electron microscopy (cryoEM), with biochemical, biophysical, computational methods. With the recent advance in direct electron detection, cryoEM has become a powerful tool for structure determination of protein complexes and assemblies. Our current research efforts are directed to two such large assemblies: HIV-1 viral capsid and bacterial chemotaxis receptor signaling arrays. In this presentation I will focus on HIV-1 capsid assembly, maturation and interaction with host cell factors that modulate viral infectivity. I will also present some of the technologies we developed, in particular the correlative fluorescent light microscopy and cryoEM method (CLEM), to advance our understanding of HIV-1 pathogenesis.
 Neocles Leontis
Neocles Leontis Karin Musier-ForsythThe 5' untranslated region (5'-UTR) is a highly conserved region of retroviral RNA genomes responsible for regulating many steps of the retroviral lifecycle including viral RNA dimerization, packaging, initiation of reverse transcription, transcriptional regulation, and splicing. A complete understanding of the mechanisms controlling retroviral replication requires structural characterization of this RNA. Unfortunately, its large size and conformational flexibility renders common methods of solving structures, such as X-ray crystallography and NMR exceedingly difficult. Here, we use a solution technique, small-angle X-ray scattering (SAXS), coupled with computational molecular modeling and structure probing, to characterize RNAs (100-350 nucleotides in length) derived from the 5'-UTR of HIV-1 (NL4-3 and MAL isolates), RSV, SIV, and HTLV-1. Similarities and differences in their packaging signals, the presence of tRNA structural mimicry, conformational switches upon dimerization and primer annealing, and length-dependent changes in global conformation will all be discussed.
Karin Musier-ForsythThe 5' untranslated region (5'-UTR) is a highly conserved region of retroviral RNA genomes responsible for regulating many steps of the retroviral lifecycle including viral RNA dimerization, packaging, initiation of reverse transcription, transcriptional regulation, and splicing. A complete understanding of the mechanisms controlling retroviral replication requires structural characterization of this RNA. Unfortunately, its large size and conformational flexibility renders common methods of solving structures, such as X-ray crystallography and NMR exceedingly difficult. Here, we use a solution technique, small-angle X-ray scattering (SAXS), coupled with computational molecular modeling and structure probing, to characterize RNAs (100-350 nucleotides in length) derived from the 5'-UTR of HIV-1 (NL4-3 and MAL isolates), RSV, SIV, and HTLV-1. Similarities and differences in their packaging signals, the presence of tRNA structural mimicry, conformational switches upon dimerization and primer annealing, and length-dependent changes in global conformation will all be discussed. Erica FlapanFor DNA molecules, topological complexity occurs exclusively as the result of knotting or linking of the polynucleotide backbone. By contrast, while a few knots and links have been found within the polypeptide backbones of some protein structures, non-planarity can also result from the connectivity between a polypeptide chain and inter- and intra-chain linking via cofactors and disulfide bonds. In this talk, we survey the known types of knots, links, and non-planar graphs in protein structures with and without including such bonds and cofactors. Then we present new examples of protein structures containing Möbius ladders and other non-planar graphs as a result of these cofactors. Finally, we propose hypothetical structures illustrating specific disulfide connectivities that would result in the key ring link, the Whitehead link and the 5-1 knot, the latter two of which have thus far not been identified within protein structures.
Erica FlapanFor DNA molecules, topological complexity occurs exclusively as the result of knotting or linking of the polynucleotide backbone. By contrast, while a few knots and links have been found within the polypeptide backbones of some protein structures, non-planarity can also result from the connectivity between a polypeptide chain and inter- and intra-chain linking via cofactors and disulfide bonds. In this talk, we survey the known types of knots, links, and non-planar graphs in protein structures with and without including such bonds and cofactors. Then we present new examples of protein structures containing Möbius ladders and other non-planar graphs as a result of these cofactors. Finally, we propose hypothetical structures illustrating specific disulfide connectivities that would result in the key ring link, the Whitehead link and the 5-1 knot, the latter two of which have thus far not been identified within protein structures. Alexander VologodskiiType II DNA topoisomerases can change DNA topology by catalyzing the passing of one double-stranded DNA segment through another. In 1997 Rybenkov et al. unexpectedly found that the enzymes can greatly reduce, up to hundred times, the fractions of knotted and linked circular DNA molecules comparing with the corresponding equilibrium values. The phenomenon of topology simplification attracted a lot of attention because it was very difficult to understand how small enzymes could determine topology of large DNA molecules. It seems clear now that the only way for the topoisomerases to achieve topology simplification is to use the fact that the probability of some specific local conformations of DNA segments depends on DNA topology. Although great progress has been made in understanding the phenomenon, some features of it are not explained by the existing models. To eliminate the discrepancy with the experimental data we suggest here a new model of the enzyme action.
Alexander VologodskiiType II DNA topoisomerases can change DNA topology by catalyzing the passing of one double-stranded DNA segment through another. In 1997 Rybenkov et al. unexpectedly found that the enzymes can greatly reduce, up to hundred times, the fractions of knotted and linked circular DNA molecules comparing with the corresponding equilibrium values. The phenomenon of topology simplification attracted a lot of attention because it was very difficult to understand how small enzymes could determine topology of large DNA molecules. It seems clear now that the only way for the topoisomerases to achieve topology simplification is to use the fact that the probability of some specific local conformations of DNA segments depends on DNA topology. Although great progress has been made in understanding the phenomenon, some features of it are not explained by the existing models. To eliminate the discrepancy with the experimental data we suggest here a new model of the enzyme action. Konstantin MischaikowNo abstract has been provided.
Konstantin MischaikowNo abstract has been provided. Mariel VazquezCellular processes such as replication, recombination, and packing change the topology of DNA. Controlling these changes is key to ensuring stability inside the cell. We use topological and computational methods to study the action of enzymes that simplify the topology of DNA during replication. I will review these methods and will expand on some thoughts on DNA folding.
Mariel VazquezCellular processes such as replication, recombination, and packing change the topology of DNA. Controlling these changes is key to ensuring stability inside the cell. We use topological and computational methods to study the action of enzymes that simplify the topology of DNA during replication. I will review these methods and will expand on some thoughts on DNA folding. Yann PontyThe prediction of the most stable, prevalent and/or functional structure adopted by an RiboNucleic Acid molecule (RNA) is an old, yet very much ongoing, challenge of computational biology. Currently available computational methods, such as MFold or RNAfold, somehow artificially restrict their search space to tree-like conformations, the secondary structures. However, such a definition intrinsically discards complex topological motifs that are both observed in experimentally-determined structures, essential for the functions performed by the molecule, and conserved throughout the evolution. In this talk, I will review two decades of works aiming at characterizing the complexity of minimizing the free energy of a given RNA molecule, while allowing (limited subsets of) pseudoknots/crossing interactions.
Yann PontyThe prediction of the most stable, prevalent and/or functional structure adopted by an RiboNucleic Acid molecule (RNA) is an old, yet very much ongoing, challenge of computational biology. Currently available computational methods, such as MFold or RNAfold, somehow artificially restrict their search space to tree-like conformations, the secondary structures. However, such a definition intrinsically discards complex topological motifs that are both observed in experimentally-determined structures, essential for the functions performed by the molecule, and conserved throughout the evolution. In this talk, I will review two decades of works aiming at characterizing the complexity of minimizing the free energy of a given RNA molecule, while allowing (limited subsets of) pseudoknots/crossing interactions. David MurrugarraBoolean networks are an important class of computational models for molecular interaction networks. Boolean canalization, a type of hierarchical clustering of the inputs of a Boolean function, has been extensively studied in the context of network modeling where each layer of canalization adds a degree of stability in the dynamics of the network. Recently, dynamic network control approaches have been used for the design of new therapeutic interventions and for other applications such as stem cell reprogramming. This talk will discuss the role of canalization in the control of Boolean molecular networks. A method for identifying the potential control edges in the wiring diagram of a network for avoiding undesirable state transitions will be presented. The method is based on identifying appropriate input-output combinations on undesirable transitions that can be modified using the edges in the wiring diagram of the network. Moreover, a method for estimating the number of changed transitions in the state space of the system as a result of an edge deletion in the wiring diagram will be presented. These two complementary methods can help in the selection of appropriate controllers such as for minimizing the side effects resulting from an edge manipulation.
David MurrugarraBoolean networks are an important class of computational models for molecular interaction networks. Boolean canalization, a type of hierarchical clustering of the inputs of a Boolean function, has been extensively studied in the context of network modeling where each layer of canalization adds a degree of stability in the dynamics of the network. Recently, dynamic network control approaches have been used for the design of new therapeutic interventions and for other applications such as stem cell reprogramming. This talk will discuss the role of canalization in the control of Boolean molecular networks. A method for identifying the potential control edges in the wiring diagram of a network for avoiding undesirable state transitions will be presented. The method is based on identifying appropriate input-output combinations on undesirable transitions that can be modified using the edges in the wiring diagram of the network. Moreover, a method for estimating the number of changed transitions in the state space of the system as a result of an edge deletion in the wiring diagram will be presented. These two complementary methods can help in the selection of appropriate controllers such as for minimizing the side effects resulting from an edge manipulation. Yusu WangI will talk about the use of topological terrain metaphors for (biological) data visualization and analysis. I will in praticular describe two software we developed: Denali, a generic tool for visualizing tree-like structures (such as clustering trees) using topological terrain metaphors, as well as Ayla, a specialized visual analytic tool for exploring molecular simulation data. This is joint work with J. Eldridge, W. Harvey, M. Belkin, T.-P. Bremer, C. Li, I. Park, V. Pascucci and O. Ruebel.
Yusu WangI will talk about the use of topological terrain metaphors for (biological) data visualization and analysis. I will in praticular describe two software we developed: Denali, a generic tool for visualizing tree-like structures (such as clustering trees) using topological terrain metaphors, as well as Ayla, a specialized visual analytic tool for exploring molecular simulation data. This is joint work with J. Eldridge, W. Harvey, M. Belkin, T.-P. Bremer, C. Li, I. Park, V. Pascucci and O. Ruebel. Robert EisenbergChemical reactions are described and analyzed using conservation of mass and the law of mass action. The conservation of mass does not imply the conservation of electric current, as can easily be seen by in the reaction A •••• B •••• C where IAB ≠IBC . The two reactions involve different rate constants, that are customarily independent, so the currents cannot be equal under more than one condition! Electric forces are very very much stronger than diffusion forces: one percent change in net charge produces a force large enough to lift the earth; one per cent change in mass has hardly any effect. I argue that chemical models cannot transferable (with one set of parameters) if they do not satisfy conservation of current. I argue that conservation of current must be exact in models of chemical reactions in all conditions, locations, and times because the ‘current’ defined in Maxwell’s equations cannot be stored, at all. My colleagues and I are trying to construct such models, following the lead of colleagues in semiconductor and computational electronics, who have done this for years.
Robert EisenbergChemical reactions are described and analyzed using conservation of mass and the law of mass action. The conservation of mass does not imply the conservation of electric current, as can easily be seen by in the reaction A •••• B •••• C where IAB ≠IBC . The two reactions involve different rate constants, that are customarily independent, so the currents cannot be equal under more than one condition! Electric forces are very very much stronger than diffusion forces: one percent change in net charge produces a force large enough to lift the earth; one per cent change in mass has hardly any effect. I argue that chemical models cannot transferable (with one set of parameters) if they do not satisfy conservation of current. I argue that conservation of current must be exact in models of chemical reactions in all conditions, locations, and times because the ‘current’ defined in Maxwell’s equations cannot be stored, at all. My colleagues and I are trying to construct such models, following the lead of colleagues in semiconductor and computational electronics, who have done this for years. Douglas TurnerLess than 5% of the human genome codes for protein, but about 90% codes for transcribed RNA. Structures and functions for much of this RNA are not known. Moreover, it appears to be more difficult to determine and to predict structures of RNAs than structures of proteins. For proteins and RNA, 3D predictions are made by both "knowledge based" and "quantum mechanical based" force fields. NMR experiments on simple RNA systems can provide benchmarks for testing methods to predict 3D structure. Results from NMR experiments and molecular dynamics simulations will be presented for single stranded, unpaired tetramers and for base paired structures of RNA. Comparisons reveal strengths and weaknesses of force fields.
Douglas TurnerLess than 5% of the human genome codes for protein, but about 90% codes for transcribed RNA. Structures and functions for much of this RNA are not known. Moreover, it appears to be more difficult to determine and to predict structures of RNAs than structures of proteins. For proteins and RNA, 3D predictions are made by both "knowledge based" and "quantum mechanical based" force fields. NMR experiments on simple RNA systems can provide benchmarks for testing methods to predict 3D structure. Results from NMR experiments and molecular dynamics simulations will be presented for single stranded, unpaired tetramers and for base paired structures of RNA. Comparisons reveal strengths and weaknesses of force fields. Alexander GrosbergIn the talk, I plan to review the current status of the controversial idea that knots in a macromolecule tighten themselves for entropic reasons.
Alexander GrosbergIn the talk, I plan to review the current status of the controversial idea that knots in a macromolecule tighten themselves for entropic reasons. Kelin XiaProteins are the most important biomolecules for living organisms. The understanding of protein structure, function, dynamics, and transport is one of the most challenging tasks in biological science. We have introduced persistent homology for extracting molecular topological fingerprints (MTFs) based on the persistence of molecular topological invariants. MTFs are utilized for protein characterization, identification, and classification. Both all-atom and coarse-grained representations of MTFs are constructed. On the basis of the correlation between protein compactness, rigidity, and connectivity, we propose an accumulated bar length generated from persistent topological invariants for the quantitative modeling of protein flexibility. To this end, a correlation matrix-based filtration is developed. This approach gives rise to an accurate prediction of the optimal characteristic distance used in protein B-factor analysis. Finally, MTFs are employed to characterize protein topological evolution during protein folding and quantitatively predict the protein folding stability. An excellent consistence between our persistent homology prediction and molecular dynamics simulation is found. This work reveals the topology-function relationship of proteins.
Kelin XiaProteins are the most important biomolecules for living organisms. The understanding of protein structure, function, dynamics, and transport is one of the most challenging tasks in biological science. We have introduced persistent homology for extracting molecular topological fingerprints (MTFs) based on the persistence of molecular topological invariants. MTFs are utilized for protein characterization, identification, and classification. Both all-atom and coarse-grained representations of MTFs are constructed. On the basis of the correlation between protein compactness, rigidity, and connectivity, we propose an accumulated bar length generated from persistent topological invariants for the quantitative modeling of protein flexibility. To this end, a correlation matrix-based filtration is developed. This approach gives rise to an accurate prediction of the optimal characteristic distance used in protein B-factor analysis. Finally, MTFs are employed to characterize protein topological evolution during protein folding and quantitatively predict the protein folding stability. An excellent consistence between our persistent homology prediction and molecular dynamics simulation is found. This work reveals the topology-function relationship of proteins. Miranda Holmes-CerfonParticles in soft-matter systems (such as colloids) tend to have very short-range interactions, so traditional theories, that assume the energy landscape is smooth enough, will struggle to capture their dynamics. We propose a new framework to look at such particles, based on taking the limit as the range of the interaction goes to zero. In this limit, the energy landscape is a set of geometrical manifolds plus a single control parameter, while the dynamics on top of the manifolds are given by a hierarchy of Fokker-Planck equations coupled by "sticky" boundary conditions. We show how to compute dynamical quantities such as transition rates between clusters of hard spheres, and then show this agrees quantitatively with experiments on colloids. We hope this framework is useful for modelling other systems with geometrical constraints, such as those that arise in biology.
Miranda Holmes-CerfonParticles in soft-matter systems (such as colloids) tend to have very short-range interactions, so traditional theories, that assume the energy landscape is smooth enough, will struggle to capture their dynamics. We propose a new framework to look at such particles, based on taking the limit as the range of the interaction goes to zero. In this limit, the energy landscape is a set of geometrical manifolds plus a single control parameter, while the dynamics on top of the manifolds are given by a hierarchy of Fokker-Planck equations coupled by "sticky" boundary conditions. We show how to compute dynamical quantities such as transition rates between clusters of hard spheres, and then show this agrees quantitatively with experiments on colloids. We hope this framework is useful for modelling other systems with geometrical constraints, such as those that arise in biology. Alan ReinRecombinant HIV-1 Gag protein, purified from bacteria, is a soluble protein, but assembles into virus-like particles (VLPs) in a defined in vitro system upon addition of nucleic acid (NA). We have tried to understand how NA-binding promotes assembly. Experiments with Gag-leucine zipper chimeric proteins suggest that when Gag attains a high local concentration, it undergoes a change which primes it for assembly; cooperative NA-binding would be one way it could reach a high local concentration. We have focused on the SP1 region of Gag, which lies between the CA domain (principally responsible for Gag-Gag interactions in VLPs) and the NC domain (principally responsible for NA-binding). Many subtle mutations in SP1 cause drastic disruption of VLP assembly. If the SP1 sequence were folded into an α-helix, the helix would be amphipathic, with a polar face and a hydrophobic face. We have found that a peptide representing SP1 folds into a helix when it is at high concentration; presumably, at these concentrations, helices can aggregate and bury their hydrophobic faces within helix bundles.
Alan ReinRecombinant HIV-1 Gag protein, purified from bacteria, is a soluble protein, but assembles into virus-like particles (VLPs) in a defined in vitro system upon addition of nucleic acid (NA). We have tried to understand how NA-binding promotes assembly. Experiments with Gag-leucine zipper chimeric proteins suggest that when Gag attains a high local concentration, it undergoes a change which primes it for assembly; cooperative NA-binding would be one way it could reach a high local concentration. We have focused on the SP1 region of Gag, which lies between the CA domain (principally responsible for Gag-Gag interactions in VLPs) and the NC domain (principally responsible for NA-binding). Many subtle mutations in SP1 cause drastic disruption of VLP assembly. If the SP1 sequence were folded into an α-helix, the helix would be amphipathic, with a polar face and a hydrophobic face. We have found that a peptide representing SP1 folds into a helix when it is at high concentration; presumably, at these concentrations, helices can aggregate and bury their hydrophobic faces within helix bundles. Peijun ZhangMy research program is interested in understanding the structural mechanisms of macromlecular assemblies using an integrated approach by combining three-dimensional cryo-electron microscopy (cryoEM), with biochemical, biophysical, computational methods. With the recent advance in direct electron detection, cryoEM has become a powerful tool for structure determination of protein complexes and assemblies. Our current research efforts are directed to two such large assemblies: HIV-1 viral capsid and bacterial chemotaxis receptor signaling arrays. In this presentation I will focus on HIV-1 capsid assembly, maturation and interaction with host cell factors that modulate viral infectivity. I will also present some of the technologies we developed, in particular the correlative fluorescent light microscopy and cryoEM method (CLEM), to advance our understanding of HIV-1 pathogenesis.
Peijun ZhangMy research program is interested in understanding the structural mechanisms of macromlecular assemblies using an integrated approach by combining three-dimensional cryo-electron microscopy (cryoEM), with biochemical, biophysical, computational methods. With the recent advance in direct electron detection, cryoEM has become a powerful tool for structure determination of protein complexes and assemblies. Our current research efforts are directed to two such large assemblies: HIV-1 viral capsid and bacterial chemotaxis receptor signaling arrays. In this presentation I will focus on HIV-1 capsid assembly, maturation and interaction with host cell factors that modulate viral infectivity. I will also present some of the technologies we developed, in particular the correlative fluorescent light microscopy and cryoEM method (CLEM), to advance our understanding of HIV-1 pathogenesis.