-

Julie Mitchell
No abstract has been provided.
-

Sharon Hammes-Schiffer
Electrostatic interactions play an important role in enzyme catalysis. These effects are modulated by the conformational changes that occur over the catalytic cycle. To elucidate the catalytic roles of these effects, thiocyanate probes were introduced at site-specific positions in the enzymes ketosteroid isomerase (KSI) and dihydrofolate reductase (DHFR). In KSI, the impact of electrostatics on ligand binding was explored. In DHFR, the impact of electrostatics on the catalytic cycle involving five different complexes was investigated. The shifts in the vibrational stretching frequencies of the nitrile probes report on the electrostatics of the microenvironments surrounding the probes. Mixed quantum mechanical/molecular mechanical molecular dynamics simulations reproduced the experimental vibrational frequency shifts and provided atomic-level insight into the roles that key residues play in determining the electrostatics of the microenvironments. The electrostatic contributions were decomposed into the major components from individual residues, ligands, and water molecules. For DHFR, calculation of the electric field along the hydride donor-acceptor axis, along with decomposition of this field into specific contributions, indicates that the cofactor and substrate, as well as the enzyme, impose a substantial electric field that facilitates hydride transfer. Overall, experimental and theoretical data provide evidence for significant electrostatic changes in the active site microenvironments due to conformational motions occurring over the catalytic cycles of enzymes.
-

Teresa Head-Gordon
Abstract not submitted.
-
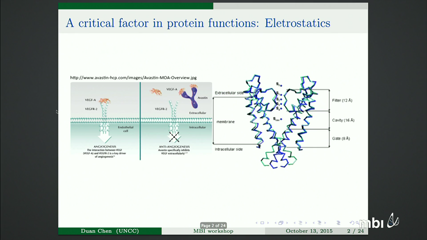
Duan Chen
Description of inhomogeneous dielectric properties of a solvent in the vicinity of ions has been attracting research interests in mathematical modeling for many years. From many experimental results, it has been concluded that the dielectric response of a solvent linearly depends on the ionic strength within a certain range. Based on this assumption, a new implicit solvent model is proposed in the form of total free energy functional and a quasi-linear Poisson-Boltzmann equation (QPBE) is derived. Classical Newton’s iteration can be used to solve the QPBE numerically but the corresponding Jacobian matrix is complicated due to the quasi-linear term. In the current work, a systematic formulation of the Jacobian matrix is derived. As an alternative option, an algorithm mixing the Newton’s iteration and the fixed point method is proposed to avoid the complicated Jacobian matrix, and it is a more general algorithm for equation with discontinuous coefficients. Computational efficiency and accuracy for these two methods are investigated based on a set of equation parameters. At last, the QPBE with singular charge source and piece-wisely defined dielectric functions has been applied to analyze electrostatics of macro biomolecules in a complicated solvent. A set of computational algorithms such as interface method, singular charge removal technique and the Newton- fixed-point iteration are employed to solve the QPBE. Biological applications of the proposed model and algorithms are provided, including calculation of electrostatic solvation free energy of proteins, investigation of physical properties of channel pore of an ion channel, and electrostatics analysis for the segment of a DNA strand.
-
Xueyu Song
In this presentation a molecular Debye-Huckel theory for ionic fluids is developed. Starting from the macroscopic Maxwell equations for bulk systems, the dispersion relation leads to a generalized Debye-Huckel theory which is related to the dressed ion theory in the static case.Due to the multi-pole structure of the dielectric function of ionic fluids, the electric potential around a single solute has a multi-Yukawa form. Given the dielectric function, the multi-Yukawa potential can be determined from our molecular Debye-Huckel theory, hence, the electrostatic contributions to thermodynamic properties of ionic fluids can be obtained. Applications to solutes with arbitrary shapes in model electrolyte solutions demonstrated the accuracy of our approach. In combination with cavity formation energy a variational approach can be used to define the boundary between a solute and its continuum solvent. Thus, our approach provides a simple and efficient path to go beyond the traditional Poisson-Boltzmann model in biophysics.
-
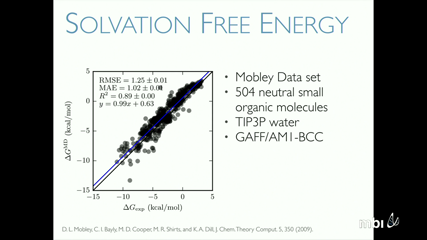
Robert Krasny
Electrostatic effects play an important role in determining protein structure and function. Here we present a treecode-accelerated boundary integral (TABI) solver for the electrostatic potential of a solvated protein described by the linear Poisson-Boltzmann equation. In this model the solvent is a continuum dielectric material with screening due to dissolved ions and the protein is a set of charged particles. The method employs a well-conditioned boundary integral formulation for the electrostatic potential and its normal derivative on the molecular surface. The surface is triangulated by MSMS and the integral equations are discretized by centroid collocation. The linear system is solved by GMRES and the matrix-vector product is carried out by a tree code which reduces the computational cost from $O(N^2)$ to $O(Nlog N)$, where $N$ is the number of faces in the triangulated molecular surface. We compare TABI results to those obtained using the finite-difference APBS code. The TABI solver exhibits good serial and parallel performance, with relatively simple implementation, efficient memory usage, and geometric adaptability. This is joint work with Weihua Geng (Southern Methodist University).
-
Tyler Luchko
No abstract has been provided.
-

Chia-en Chang
No abstract has been provided.
-
Zhan Chen
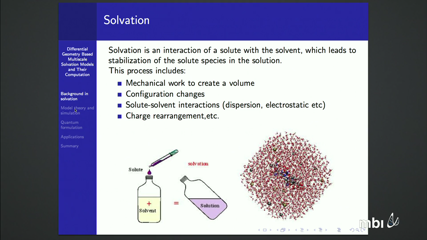
The understanding of solvation is an essential prerequisite for the quantitative description and analysis of many sophisticated chemical, biological and biomolecular processes.. Implicit solvent models, particularly those based on the Poisson-Boltzmann (PB) equation for electrostatic analysis, are established approaches for solvation analysis. However, ad hoc solvent-solute interfaces and complex solutions of nonlinear equations post some severe limitations on their applications.
We have introduced differential geometry (DG) based multiscale solvation models, which allow the solvent-solute interface, electrostatic potential, and even electron densities to be determined by the variation of a total free energy functional. In addition, our models is able to significantly reduce the number of free parameters and to avoid complicated interface problems raised by sharp solvent-solute interface. Finally, our DG based models have shown promising power in blind prediction of solvation as well as other applications. This is primarily joint work with Prof. Guowei Wei and Prof. Nathan Baker.
-

Marharyta Petukh
Ions play significant role in all living cells. They are involved in multiple biological processes by maintaining the unique fold of macromolecules; participating in their enzymatic activity; and screening electrostatic interactions. While experimental methods are not always able to assign the exact location of ions, computational methods are in demand. Although the majority of computational methods are successful in predicting the position of ions buried inside macromolecules, they are less effective in deciphering positions of surface bound ions. The new BION algorithm (compbio.clemson.edu/bion_server_pH/) predicts the location of ions of the surface of proteins based on electrostatic and geometrical properties of both ions and proteins. The advanced clustering procedure in combination with pairing rules improves both the efficiency and the accuracy of the method. The webserver allows specifying the pH and the salt concentration in predicting ions positions. The example of BION application is demonstrated in the study of the origin of the Chronic Berillium Disease and functionality of TAR/TSR bacterial chemoreceptors.
-
Lin Li
One of the most important roles of Kinesins is transporting cargo and organelles along microtubules. N-kinesins proceeding toward the plus-end of microtubule while C-kinesin proceeding toward the minus end. Many experimental researches and few computational efforts have been performed to investigate the mechanism of kinesins’ proceeding. However, many details of the mechanism for the kinesins’ proceeding are still unknown. One of the reason is the size of the kinesin-microtubule system is too large for simulations. A Monte Carlo simulation approach with discrete conformational sampling algorithm is developed to reveal the role of electrostatic interactions in kinesins’ proceeding on microtubule. We find that the electrostatic profile on microtubule forms potential valleys around the microtubule, which keeps kinesin walking along the longitudinal direction of microtubule rather than lateral direction. The simulated pathways on N-kinesin, C-kinesin and nanoparticle show that the electrostatic interactions guide the kinesins walking toward their directions.
-

Ridgway Scott
Dipoles are ubiquitous in nature. Many materials are made of dipolar molecules, such as water. Thus it is of interest to know how large collections of dipoles can interact on a macro scale. One measure of this is called the Madelung constant. Materials whose dipoles coordinate on a global scale are called ferro-electric, by analogy with ferro-magnets. Ferro-electric materials can store a permanent charge. We describe how it is possible for water ice to become ferro-electric, and we discuss how to interpret Madelung constants in cases where the corresponding sum of dipoles appears divergent.
-

Donald Jacobs
Proteins are macromolecules consisting of myriad intramolecular interactions together with interactions with solvent that determine their conformational ensemble, stability and dynamics. Global constraints such as temperature and solvent composition play an essential role in defining equilibrium properties. Similarly, covalent bonding and other intramolecular interactions such as hydrogen bonds impose local mechanical constraints. Application of graph-rigidity has played an important role in predicting protein flexibility, exploring conformational dynamics through geometrical simulation (GS) and predicting thermodynamic stability via a Distance Constraint Model (DCM) that accounts for non-additivity in conformational entropy. A DCM/GS hybrid method is presented that rapidly explores conformational dynamics guided by changes in free energy by successively solving a free energy functional. A critical part of the free energy functional is modeling the solvation contribution to balance accuracy and speed so as to enable rapid exploration of conformational space that is scalable to a collection of proteins in a multi-component solvent to investigate protein-protein interactions in specific formulations and the cellular environment. Among the many implicit solvation models available in the literature, two approaches are being pursued currently which will be used to generate discussions among the experts.
-

Nathan Baker
Biomolecules exhibit conformational fluctuations near equilibrium states, inducing uncertainty in various biological properties in a dynamic way. We have developed a general method to quantify the uncertainty of target properties induced by conformational fluctuations. Using a generalized polynomial chaos (gPC) expansion, we construct a surrogate model of the target property with respect to varying conformational states. We also propose a method to increase the sparsity of the gPC expansion by defining a set of conformational "active space" random variables. With the increased sparsity, we employ the compressive sensing method to accurately construct the surrogate model. We demonstrate the performance of the surrogate model by evaluating fluctuation-induced uncertainty in solvent-accessible surface area for the bovine trypsin inhibitor protein system and show that the new approach offers more accurate statistical information than standard Monte Carlo approaches. Further more, the constructed surrogate model also enables us to directly evaluate the target property under various conformational states, yielding a more accurate response surface than standard sparse grid collocation methods. In particular, the new method provides higher accuracy in high-dimensional systems, such as biomolecules, where sparse grid performance is limited by the accuracy of the computed quantity of interest. Our new framework is generalizable and can be used to investigate the uncertainty of a wide variety of target properties in biomolecular systems.
 Julie MitchellNo abstract has been provided.
Julie MitchellNo abstract has been provided. Sharon Hammes-SchifferElectrostatic interactions play an important role in enzyme catalysis. These effects are modulated by the conformational changes that occur over the catalytic cycle. To elucidate the catalytic roles of these effects, thiocyanate probes were introduced at site-specific positions in the enzymes ketosteroid isomerase (KSI) and dihydrofolate reductase (DHFR). In KSI, the impact of electrostatics on ligand binding was explored. In DHFR, the impact of electrostatics on the catalytic cycle involving five different complexes was investigated. The shifts in the vibrational stretching frequencies of the nitrile probes report on the electrostatics of the microenvironments surrounding the probes. Mixed quantum mechanical/molecular mechanical molecular dynamics simulations reproduced the experimental vibrational frequency shifts and provided atomic-level insight into the roles that key residues play in determining the electrostatics of the microenvironments. The electrostatic contributions were decomposed into the major components from individual residues, ligands, and water molecules. For DHFR, calculation of the electric field along the hydride donor-acceptor axis, along with decomposition of this field into specific contributions, indicates that the cofactor and substrate, as well as the enzyme, impose a substantial electric field that facilitates hydride transfer. Overall, experimental and theoretical data provide evidence for significant electrostatic changes in the active site microenvironments due to conformational motions occurring over the catalytic cycles of enzymes.
Sharon Hammes-SchifferElectrostatic interactions play an important role in enzyme catalysis. These effects are modulated by the conformational changes that occur over the catalytic cycle. To elucidate the catalytic roles of these effects, thiocyanate probes were introduced at site-specific positions in the enzymes ketosteroid isomerase (KSI) and dihydrofolate reductase (DHFR). In KSI, the impact of electrostatics on ligand binding was explored. In DHFR, the impact of electrostatics on the catalytic cycle involving five different complexes was investigated. The shifts in the vibrational stretching frequencies of the nitrile probes report on the electrostatics of the microenvironments surrounding the probes. Mixed quantum mechanical/molecular mechanical molecular dynamics simulations reproduced the experimental vibrational frequency shifts and provided atomic-level insight into the roles that key residues play in determining the electrostatics of the microenvironments. The electrostatic contributions were decomposed into the major components from individual residues, ligands, and water molecules. For DHFR, calculation of the electric field along the hydride donor-acceptor axis, along with decomposition of this field into specific contributions, indicates that the cofactor and substrate, as well as the enzyme, impose a substantial electric field that facilitates hydride transfer. Overall, experimental and theoretical data provide evidence for significant electrostatic changes in the active site microenvironments due to conformational motions occurring over the catalytic cycles of enzymes. Teresa Head-GordonAbstract not submitted.
Teresa Head-GordonAbstract not submitted. Duan ChenDescription of inhomogeneous dielectric properties of a solvent in the vicinity of ions has been attracting research interests in mathematical modeling for many years. From many experimental results, it has been concluded that the dielectric response of a solvent linearly depends on the ionic strength within a certain range. Based on this assumption, a new implicit solvent model is proposed in the form of total free energy functional and a quasi-linear Poisson-Boltzmann equation (QPBE) is derived. Classical Newton’s iteration can be used to solve the QPBE numerically but the corresponding Jacobian matrix is complicated due to the quasi-linear term. In the current work, a systematic formulation of the Jacobian matrix is derived. As an alternative option, an algorithm mixing the Newton’s iteration and the fixed point method is proposed to avoid the complicated Jacobian matrix, and it is a more general algorithm for equation with discontinuous coefficients. Computational efficiency and accuracy for these two methods are investigated based on a set of equation parameters. At last, the QPBE with singular charge source and piece-wisely defined dielectric functions has been applied to analyze electrostatics of macro biomolecules in a complicated solvent. A set of computational algorithms such as interface method, singular charge removal technique and the Newton- fixed-point iteration are employed to solve the QPBE. Biological applications of the proposed model and algorithms are provided, including calculation of electrostatic solvation free energy of proteins, investigation of physical properties of channel pore of an ion channel, and electrostatics analysis for the segment of a DNA strand.
Duan ChenDescription of inhomogeneous dielectric properties of a solvent in the vicinity of ions has been attracting research interests in mathematical modeling for many years. From many experimental results, it has been concluded that the dielectric response of a solvent linearly depends on the ionic strength within a certain range. Based on this assumption, a new implicit solvent model is proposed in the form of total free energy functional and a quasi-linear Poisson-Boltzmann equation (QPBE) is derived. Classical Newton’s iteration can be used to solve the QPBE numerically but the corresponding Jacobian matrix is complicated due to the quasi-linear term. In the current work, a systematic formulation of the Jacobian matrix is derived. As an alternative option, an algorithm mixing the Newton’s iteration and the fixed point method is proposed to avoid the complicated Jacobian matrix, and it is a more general algorithm for equation with discontinuous coefficients. Computational efficiency and accuracy for these two methods are investigated based on a set of equation parameters. At last, the QPBE with singular charge source and piece-wisely defined dielectric functions has been applied to analyze electrostatics of macro biomolecules in a complicated solvent. A set of computational algorithms such as interface method, singular charge removal technique and the Newton- fixed-point iteration are employed to solve the QPBE. Biological applications of the proposed model and algorithms are provided, including calculation of electrostatic solvation free energy of proteins, investigation of physical properties of channel pore of an ion channel, and electrostatics analysis for the segment of a DNA strand. Xueyu SongIn this presentation a molecular Debye-Huckel theory for ionic fluids is developed. Starting from the macroscopic Maxwell equations for bulk systems, the dispersion relation leads to a generalized Debye-Huckel theory which is related to the dressed ion theory in the static case.Due to the multi-pole structure of the dielectric function of ionic fluids, the electric potential around a single solute has a multi-Yukawa form. Given the dielectric function, the multi-Yukawa potential can be determined from our molecular Debye-Huckel theory, hence, the electrostatic contributions to thermodynamic properties of ionic fluids can be obtained. Applications to solutes with arbitrary shapes in model electrolyte solutions demonstrated the accuracy of our approach. In combination with cavity formation energy a variational approach can be used to define the boundary between a solute and its continuum solvent. Thus, our approach provides a simple and efficient path to go beyond the traditional Poisson-Boltzmann model in biophysics.
Xueyu SongIn this presentation a molecular Debye-Huckel theory for ionic fluids is developed. Starting from the macroscopic Maxwell equations for bulk systems, the dispersion relation leads to a generalized Debye-Huckel theory which is related to the dressed ion theory in the static case.Due to the multi-pole structure of the dielectric function of ionic fluids, the electric potential around a single solute has a multi-Yukawa form. Given the dielectric function, the multi-Yukawa potential can be determined from our molecular Debye-Huckel theory, hence, the electrostatic contributions to thermodynamic properties of ionic fluids can be obtained. Applications to solutes with arbitrary shapes in model electrolyte solutions demonstrated the accuracy of our approach. In combination with cavity formation energy a variational approach can be used to define the boundary between a solute and its continuum solvent. Thus, our approach provides a simple and efficient path to go beyond the traditional Poisson-Boltzmann model in biophysics. Robert KrasnyElectrostatic effects play an important role in determining protein structure and function. Here we present a treecode-accelerated boundary integral (TABI) solver for the electrostatic potential of a solvated protein described by the linear Poisson-Boltzmann equation. In this model the solvent is a continuum dielectric material with screening due to dissolved ions and the protein is a set of charged particles. The method employs a well-conditioned boundary integral formulation for the electrostatic potential and its normal derivative on the molecular surface. The surface is triangulated by MSMS and the integral equations are discretized by centroid collocation. The linear system is solved by GMRES and the matrix-vector product is carried out by a tree code which reduces the computational cost from $O(N^2)$ to $O(Nlog N)$, where $N$ is the number of faces in the triangulated molecular surface. We compare TABI results to those obtained using the finite-difference APBS code. The TABI solver exhibits good serial and parallel performance, with relatively simple implementation, efficient memory usage, and geometric adaptability. This is joint work with Weihua Geng (Southern Methodist University).
Robert KrasnyElectrostatic effects play an important role in determining protein structure and function. Here we present a treecode-accelerated boundary integral (TABI) solver for the electrostatic potential of a solvated protein described by the linear Poisson-Boltzmann equation. In this model the solvent is a continuum dielectric material with screening due to dissolved ions and the protein is a set of charged particles. The method employs a well-conditioned boundary integral formulation for the electrostatic potential and its normal derivative on the molecular surface. The surface is triangulated by MSMS and the integral equations are discretized by centroid collocation. The linear system is solved by GMRES and the matrix-vector product is carried out by a tree code which reduces the computational cost from $O(N^2)$ to $O(Nlog N)$, where $N$ is the number of faces in the triangulated molecular surface. We compare TABI results to those obtained using the finite-difference APBS code. The TABI solver exhibits good serial and parallel performance, with relatively simple implementation, efficient memory usage, and geometric adaptability. This is joint work with Weihua Geng (Southern Methodist University). Tyler LuchkoNo abstract has been provided.
Tyler LuchkoNo abstract has been provided. Chia-en ChangNo abstract has been provided.
Chia-en ChangNo abstract has been provided. Zhan ChenThe understanding of solvation is an essential prerequisite for the quantitative description and analysis of many sophisticated chemical, biological and biomolecular processes.. Implicit solvent models, particularly those based on the Poisson-Boltzmann (PB) equation for electrostatic analysis, are established approaches for solvation analysis. However, ad hoc solvent-solute interfaces and complex solutions of nonlinear equations post some severe limitations on their applications.
Zhan ChenThe understanding of solvation is an essential prerequisite for the quantitative description and analysis of many sophisticated chemical, biological and biomolecular processes.. Implicit solvent models, particularly those based on the Poisson-Boltzmann (PB) equation for electrostatic analysis, are established approaches for solvation analysis. However, ad hoc solvent-solute interfaces and complex solutions of nonlinear equations post some severe limitations on their applications. Marharyta PetukhIons play significant role in all living cells. They are involved in multiple biological processes by maintaining the unique fold of macromolecules; participating in their enzymatic activity; and screening electrostatic interactions. While experimental methods are not always able to assign the exact location of ions, computational methods are in demand. Although the majority of computational methods are successful in predicting the position of ions buried inside macromolecules, they are less effective in deciphering positions of surface bound ions. The new BION algorithm (compbio.clemson.edu/bion_server_pH/) predicts the location of ions of the surface of proteins based on electrostatic and geometrical properties of both ions and proteins. The advanced clustering procedure in combination with pairing rules improves both the efficiency and the accuracy of the method. The webserver allows specifying the pH and the salt concentration in predicting ions positions. The example of BION application is demonstrated in the study of the origin of the Chronic Berillium Disease and functionality of TAR/TSR bacterial chemoreceptors.
Marharyta PetukhIons play significant role in all living cells. They are involved in multiple biological processes by maintaining the unique fold of macromolecules; participating in their enzymatic activity; and screening electrostatic interactions. While experimental methods are not always able to assign the exact location of ions, computational methods are in demand. Although the majority of computational methods are successful in predicting the position of ions buried inside macromolecules, they are less effective in deciphering positions of surface bound ions. The new BION algorithm (compbio.clemson.edu/bion_server_pH/) predicts the location of ions of the surface of proteins based on electrostatic and geometrical properties of both ions and proteins. The advanced clustering procedure in combination with pairing rules improves both the efficiency and the accuracy of the method. The webserver allows specifying the pH and the salt concentration in predicting ions positions. The example of BION application is demonstrated in the study of the origin of the Chronic Berillium Disease and functionality of TAR/TSR bacterial chemoreceptors. Lin LiOne of the most important roles of Kinesins is transporting cargo and organelles along microtubules. N-kinesins proceeding toward the plus-end of microtubule while C-kinesin proceeding toward the minus end. Many experimental researches and few computational efforts have been performed to investigate the mechanism of kinesins’ proceeding. However, many details of the mechanism for the kinesins’ proceeding are still unknown. One of the reason is the size of the kinesin-microtubule system is too large for simulations. A Monte Carlo simulation approach with discrete conformational sampling algorithm is developed to reveal the role of electrostatic interactions in kinesins’ proceeding on microtubule. We find that the electrostatic profile on microtubule forms potential valleys around the microtubule, which keeps kinesin walking along the longitudinal direction of microtubule rather than lateral direction. The simulated pathways on N-kinesin, C-kinesin and nanoparticle show that the electrostatic interactions guide the kinesins walking toward their directions.
Lin LiOne of the most important roles of Kinesins is transporting cargo and organelles along microtubules. N-kinesins proceeding toward the plus-end of microtubule while C-kinesin proceeding toward the minus end. Many experimental researches and few computational efforts have been performed to investigate the mechanism of kinesins’ proceeding. However, many details of the mechanism for the kinesins’ proceeding are still unknown. One of the reason is the size of the kinesin-microtubule system is too large for simulations. A Monte Carlo simulation approach with discrete conformational sampling algorithm is developed to reveal the role of electrostatic interactions in kinesins’ proceeding on microtubule. We find that the electrostatic profile on microtubule forms potential valleys around the microtubule, which keeps kinesin walking along the longitudinal direction of microtubule rather than lateral direction. The simulated pathways on N-kinesin, C-kinesin and nanoparticle show that the electrostatic interactions guide the kinesins walking toward their directions. Ridgway ScottDipoles are ubiquitous in nature. Many materials are made of dipolar molecules, such as water. Thus it is of interest to know how large collections of dipoles can interact on a macro scale. One measure of this is called the Madelung constant. Materials whose dipoles coordinate on a global scale are called ferro-electric, by analogy with ferro-magnets. Ferro-electric materials can store a permanent charge. We describe how it is possible for water ice to become ferro-electric, and we discuss how to interpret Madelung constants in cases where the corresponding sum of dipoles appears divergent.
Ridgway ScottDipoles are ubiquitous in nature. Many materials are made of dipolar molecules, such as water. Thus it is of interest to know how large collections of dipoles can interact on a macro scale. One measure of this is called the Madelung constant. Materials whose dipoles coordinate on a global scale are called ferro-electric, by analogy with ferro-magnets. Ferro-electric materials can store a permanent charge. We describe how it is possible for water ice to become ferro-electric, and we discuss how to interpret Madelung constants in cases where the corresponding sum of dipoles appears divergent. Donald JacobsProteins are macromolecules consisting of myriad intramolecular interactions together with interactions with solvent that determine their conformational ensemble, stability and dynamics. Global constraints such as temperature and solvent composition play an essential role in defining equilibrium properties. Similarly, covalent bonding and other intramolecular interactions such as hydrogen bonds impose local mechanical constraints. Application of graph-rigidity has played an important role in predicting protein flexibility, exploring conformational dynamics through geometrical simulation (GS) and predicting thermodynamic stability via a Distance Constraint Model (DCM) that accounts for non-additivity in conformational entropy. A DCM/GS hybrid method is presented that rapidly explores conformational dynamics guided by changes in free energy by successively solving a free energy functional. A critical part of the free energy functional is modeling the solvation contribution to balance accuracy and speed so as to enable rapid exploration of conformational space that is scalable to a collection of proteins in a multi-component solvent to investigate protein-protein interactions in specific formulations and the cellular environment. Among the many implicit solvation models available in the literature, two approaches are being pursued currently which will be used to generate discussions among the experts.
Donald JacobsProteins are macromolecules consisting of myriad intramolecular interactions together with interactions with solvent that determine their conformational ensemble, stability and dynamics. Global constraints such as temperature and solvent composition play an essential role in defining equilibrium properties. Similarly, covalent bonding and other intramolecular interactions such as hydrogen bonds impose local mechanical constraints. Application of graph-rigidity has played an important role in predicting protein flexibility, exploring conformational dynamics through geometrical simulation (GS) and predicting thermodynamic stability via a Distance Constraint Model (DCM) that accounts for non-additivity in conformational entropy. A DCM/GS hybrid method is presented that rapidly explores conformational dynamics guided by changes in free energy by successively solving a free energy functional. A critical part of the free energy functional is modeling the solvation contribution to balance accuracy and speed so as to enable rapid exploration of conformational space that is scalable to a collection of proteins in a multi-component solvent to investigate protein-protein interactions in specific formulations and the cellular environment. Among the many implicit solvation models available in the literature, two approaches are being pursued currently which will be used to generate discussions among the experts. Nathan BakerBiomolecules exhibit conformational fluctuations near equilibrium states, inducing uncertainty in various biological properties in a dynamic way. We have developed a general method to quantify the uncertainty of target properties induced by conformational fluctuations. Using a generalized polynomial chaos (gPC) expansion, we construct a surrogate model of the target property with respect to varying conformational states. We also propose a method to increase the sparsity of the gPC expansion by defining a set of conformational "active space" random variables. With the increased sparsity, we employ the compressive sensing method to accurately construct the surrogate model. We demonstrate the performance of the surrogate model by evaluating fluctuation-induced uncertainty in solvent-accessible surface area for the bovine trypsin inhibitor protein system and show that the new approach offers more accurate statistical information than standard Monte Carlo approaches. Further more, the constructed surrogate model also enables us to directly evaluate the target property under various conformational states, yielding a more accurate response surface than standard sparse grid collocation methods. In particular, the new method provides higher accuracy in high-dimensional systems, such as biomolecules, where sparse grid performance is limited by the accuracy of the computed quantity of interest. Our new framework is generalizable and can be used to investigate the uncertainty of a wide variety of target properties in biomolecular systems.
Nathan BakerBiomolecules exhibit conformational fluctuations near equilibrium states, inducing uncertainty in various biological properties in a dynamic way. We have developed a general method to quantify the uncertainty of target properties induced by conformational fluctuations. Using a generalized polynomial chaos (gPC) expansion, we construct a surrogate model of the target property with respect to varying conformational states. We also propose a method to increase the sparsity of the gPC expansion by defining a set of conformational "active space" random variables. With the increased sparsity, we employ the compressive sensing method to accurately construct the surrogate model. We demonstrate the performance of the surrogate model by evaluating fluctuation-induced uncertainty in solvent-accessible surface area for the bovine trypsin inhibitor protein system and show that the new approach offers more accurate statistical information than standard Monte Carlo approaches. Further more, the constructed surrogate model also enables us to directly evaluate the target property under various conformational states, yielding a more accurate response surface than standard sparse grid collocation methods. In particular, the new method provides higher accuracy in high-dimensional systems, such as biomolecules, where sparse grid performance is limited by the accuracy of the computed quantity of interest. Our new framework is generalizable and can be used to investigate the uncertainty of a wide variety of target properties in biomolecular systems.