-
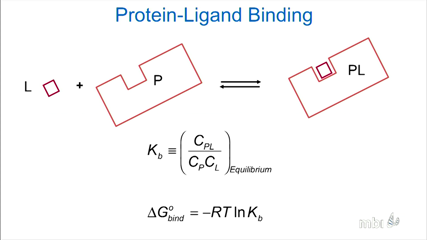
Michael Gilson
-

Huan Lei
-
Emil Alexov
DNA mutations are the cause of many human diseases and they are also the reason for natural differences among individuals by affecting the structure, function, interactions, and other properties of DNA and expressed proteins. Some diseases are caused by mutations in several genes, while others are caused by defects in a single gene (monogenic diseases). Here we focus on two monogenic diseases: (a) the Snyder-Robinson Syndrome (SRS) which is a rare mental retardation disorder caused by missense mutations in spermine synthase (SMS) and (b) the Rett syndrome (RTT) which is a brain disorder that is linked with mutations in MeCP2 protein, and it is estimated to affect 1 in 8,500 females. We demonstrate that the vast majority of mutations do not directly affect the functionality of the corresponding protein, but rather alter its stability and affinity. This prompted us to seek small molecules which target the mutant protein and upon the binding restore its wild type characteristics. The computational findings are experimentally verified.
-

Dexuan Xie
Calculation of electrostatics for a biomolecule (or a complex of a protein with a drug molecule) in an ionic solvent is a fundamental task in the fields of structural biology, computational biochemistry, biophysics, and mathematical biology. The Poisson-Boltzmann equation (PBE) is one commonly used dielectric model for predicting electrostatics of ionic solvated biomolecules. It has played important roles in rational drug design and protein design as well as other bioengineering applications. However, it is known not to work properly near a highly charged biomolecular surface, since it does not reflect any polarization correlation among water molecules and ionic size effects.
To improve the quality of PBE in the calculation of electrostatic solvation and binding free energies, we made many progresses recently on the study of nonlocal dielectric models, and developed several fast nonlocal model solvers. Meanwhile, we developed new numerical algorithms for solving PBE and one size modified PBE by using finite element, finite difference, solution decomposition, domain decomposition, and multigrid methods.
In this talk, I will first review our nonlocal dielectric theory. I then will present a new nonlocal PBE and its finite element solver. I will also describe our new numerical algorithms for solving PBE and one size modified PBE. A collection of these new solvers has led to a new software tool, called SDPBS (Solution Decomposition Poisson-Boltzmann Solvers), which is available online for free through our web server. Finally, application examples for chemical molecules, proteins, protein-drug, and peptide-RNA will be given to demonstrate the high performance and numerical stability of SDPBE in the calculation of salvation and binding free energies. This project is a joined work with Prof. L. Ridgway Scott at the University of Chicago under the support by NSF grants (DMS-0921004, DMS-1226259, and DMS-1226019).
-
Christopher Langmead
-

Tony Lelievre
I will present a numerical method that we are currently developing in the NAMD software and which aim at simulation reactive trajectories. Numerical and theoretical results on this technique will be described.
-
Alex MacKerell
Computational functional group affinity mapping of proteins is of utility for ligand design in the context of database screening, fragment-based design and lead compound optimization. The SILCS methodology allows for the generation of functional group affinity maps (FragMaps) of proteins that take into account contributions from protein desolvation, functional group desolvation, protein flexibility as well as direct interactions of the functional groups with the protein. Boltzmann transformation of the maps yields Grid Free Energy (GFE) FragMaps that may be used both qualitatively and quantitatively to direct ligand design. To allow for the application of the SILCS approach to deep and occluded pockets in proteins an oscillating μex Grand Canonical Monte Carlo (GCMC) approach was developed that allows for insertions of small solute molecules in the presence of an explicit aqueous environment. Combining the GCMC method with MD simulations for the inclusion of protein flexibility allows for the determination of GFE Fragmaps in occluded pockets. An overview of the GCMC/MD SILCS methodology along with application of the method to T4-lysozyme pocket mutants, nuclear receptors and GPCRs will be presented.
-
Sandor Vajda
Binding energy hot spots, smaller regions of binding sites that contribute a disproportionate amount to the free energy of binding any ligand, can be determined computationally from ligand-free structures of protein targets. The hot spot structure of a protein target provides very useful information on binding properties. The first application that will be discussed is predicting druggability, i.e., the ability of a site of binding druglike ligands with sufficient affinity. We applied the method to a large set of proteins. Results showed that, because the method is based on the biophysics of binding rather than on empirical parameterization, meaningful information can be gained about classes of proteins and classes of compounds beyond those resembling validated targets and conventionally druglike ligands. In particular, the method identifies targets that, while not druggable by druglike compounds, may become druggable using compound classes such as macrocycles or other large molecules beyond the rule-of-five limit. Second, we show that the hot spots provide crucial insights into the prospects for successful application of fragment-based drug discovery (FBDD), and whether a fragment hit can be advanced into a high affinity ligand. The key factor is the strength of the top ranking hot spot, and how well a given fragment complements it. We show that published data are sufficient to provide a sophisticated and quantitative understanding of how hot spot strength, number and spatial arrangement govern the potential for a surface site to bind to fragment-sized and larger ligands. This improved understanding provides important guidance for the effective application of FBDD in drug discovery.
-

Cameron Abrams
Prediction of binding and unbinding kinetics from all-atom molecular simulations could one day be an important component in the toolkit for structure-based drug design. Unfortunately, in contrast to binding affinity prediction, algorithms for estimating on- and off-rates in relatively complicated systems remain relatively undeveloped. Here we demonstrate one approach that has been applied to successfully compute the entry and exit rates of small gas molecules into globular proteins. The method is an application of transition-path theory and involves performing milestoning MD simulations in cells forming a Voronoi tesselation of some chosen state-discriminating collective-variable space. Particular attention is paid to handshaking the simulations with a continuous approximation of the solute flux to a protein target surface as a function of ligand diffusivity and bulk concentration. The method is applicable beyond small gas molecules and extensions involving more complicated molecules will be discussed.
-

Chris Chipot
One of the grand challenges of rationale drug design is the prediction of the affinity of potential therapeutic agents for a given protein target. This challenge is in large measure rooted in the considerable changes in configurational entropy that accompanies the binding process, which atomistic simulations cannot easily sample. Two strategies relying upon alchemical transformations, on the one hand, and geometric transformations, notably potential of mean force calculations, on the other hand, are proposed, invoking a series of geometric restraints acting on collecting variables designed to alleviate sampling limitations inherent to classical molecular dynamics simulations. I will show through the example of a protein binding a small substrate, that both strategies, however of clearly different nature, can yield nearly identical standard binding free energies within chemical accuracy. I will further show how the methodology can be seamlessly transposed to protein-protein complexes. I will also outline current strategies to estimate binding entropies from such calculations. Downstream from the prediction of binding affinities is the challenging prediction of bioavailability. To estimate the permeability of the biological membrane to a drug candidate, an approach based upon Bayesian inferences, which reconciles thermodynamics and kinetics in molecular dynamics simulations with time-dependent biases, is put forth. Performance of the method is illustrated with prototypical permeants diffusing in a homogeneous lipid bilayer.
-

Martin Goethe
Minimizing a suitable free energy expression is arguably the most common approach in (ab initio) protein structure prediction. The achieved accuracy depends crucially on the quality of the free energy expression in use. Here, we present corrections to existing free energy expressions which arise from the thermal motion of the protein. We (i) devise a term accounting for the vibrational entropy of the protein, and (ii) correct existing potentials for the "thermal smoothing effect".
(i) Vibrational entropy is almost always neglected in free energy expressions as its consideration is difficult. This practice, however, may lead to incorrect output because distinct conformations of a protein can contain very different amount of vibrational entropy, as we show for the chicken villin headpiece explicitly [1]. For considering vibrational entropy, we suggest a knowledge based approach where typical fluctuation and correlation patterns are extracted from known proteins and then applied to new targets.
(ii) At ambient conditions, time-averaged potentials of proteins are considerably smoother when expressed in terms of the average atom coordinates than the Hamiltonian. This effect caused by thermal motion is referred to as the thermal smoothing effect. The strength of the effect varies strongly between atoms. This allows to increase the accuracy of free energy expressions significantly by subdividing atom species regarding their typical fluctuation behavior inside proteins and assigning time-averaged potentials for the new sub-species independently [2].
[1] M. Goethe, I. Fita, and J.M. Rubi, Vibrational Entropy of a Protein: Large Differences between Distinct Conformations, J. Chem. Theory Comput. 11, 351 (2015).
[2] M. Goethe, I. Fita, and J.M. Rubi, Thermal Motion in Proteins: Large Effects on the Time-Averaged Interaction Energies, in revision.
-
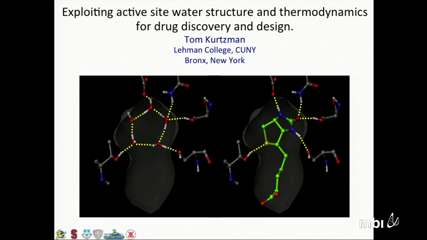
Tom Kurtzman
Despite recent advances in methodologies that better characterize local water structure and thermodynamics, the simply- stated question of whether it would be beneficial or detrimental, in a free-energetic sense, to displace water from a region of a protein with a suitably complementary ligand continues to be a conundrum. Solvation thermodynamic mapping techniques have provided valuable insight into the role of displacing water in molecular recognition however solvation thermodynamics alone is not predictive of displaceability and tightly binding ligands regularly displace water from both regions that are characterized as having favorable solvation and regions that are characterized as having unfavorable solvation. Part of the difficulty of assessing the thermodynamics of water displacement is due to the large number of often counteracting contributions and the consequent lack of a simplifying conceptual framework. We will discuss the key insights which have been gained from solvation thermodynamic mapping and how we are incorporating them into drug discovery and design methodologies. We will also discuss remaining issues on predicting favorability of water displacement and suggest how they might be tackled.
 Michael Gilson
Michael Gilson Huan Lei
Huan Lei Emil AlexovDNA mutations are the cause of many human diseases and they are also the reason for natural differences among individuals by affecting the structure, function, interactions, and other properties of DNA and expressed proteins. Some diseases are caused by mutations in several genes, while others are caused by defects in a single gene (monogenic diseases). Here we focus on two monogenic diseases: (a) the Snyder-Robinson Syndrome (SRS) which is a rare mental retardation disorder caused by missense mutations in spermine synthase (SMS) and (b) the Rett syndrome (RTT) which is a brain disorder that is linked with mutations in MeCP2 protein, and it is estimated to affect 1 in 8,500 females. We demonstrate that the vast majority of mutations do not directly affect the functionality of the corresponding protein, but rather alter its stability and affinity. This prompted us to seek small molecules which target the mutant protein and upon the binding restore its wild type characteristics. The computational findings are experimentally verified.
Emil AlexovDNA mutations are the cause of many human diseases and they are also the reason for natural differences among individuals by affecting the structure, function, interactions, and other properties of DNA and expressed proteins. Some diseases are caused by mutations in several genes, while others are caused by defects in a single gene (monogenic diseases). Here we focus on two monogenic diseases: (a) the Snyder-Robinson Syndrome (SRS) which is a rare mental retardation disorder caused by missense mutations in spermine synthase (SMS) and (b) the Rett syndrome (RTT) which is a brain disorder that is linked with mutations in MeCP2 protein, and it is estimated to affect 1 in 8,500 females. We demonstrate that the vast majority of mutations do not directly affect the functionality of the corresponding protein, but rather alter its stability and affinity. This prompted us to seek small molecules which target the mutant protein and upon the binding restore its wild type characteristics. The computational findings are experimentally verified. Dexuan XieCalculation of electrostatics for a biomolecule (or a complex of a protein with a drug molecule) in an ionic solvent is a fundamental task in the fields of structural biology, computational biochemistry, biophysics, and mathematical biology. The Poisson-Boltzmann equation (PBE) is one commonly used dielectric model for predicting electrostatics of ionic solvated biomolecules. It has played important roles in rational drug design and protein design as well as other bioengineering applications. However, it is known not to work properly near a highly charged biomolecular surface, since it does not reflect any polarization correlation among water molecules and ionic size effects.
Dexuan XieCalculation of electrostatics for a biomolecule (or a complex of a protein with a drug molecule) in an ionic solvent is a fundamental task in the fields of structural biology, computational biochemistry, biophysics, and mathematical biology. The Poisson-Boltzmann equation (PBE) is one commonly used dielectric model for predicting electrostatics of ionic solvated biomolecules. It has played important roles in rational drug design and protein design as well as other bioengineering applications. However, it is known not to work properly near a highly charged biomolecular surface, since it does not reflect any polarization correlation among water molecules and ionic size effects. Christopher Langmead
Christopher Langmead Tony LelievreI will present a numerical method that we are currently developing in the NAMD software and which aim at simulation reactive trajectories. Numerical and theoretical results on this technique will be described.
Tony LelievreI will present a numerical method that we are currently developing in the NAMD software and which aim at simulation reactive trajectories. Numerical and theoretical results on this technique will be described. Alex MacKerellComputational functional group affinity mapping of proteins is of utility for ligand design in the context of database screening, fragment-based design and lead compound optimization. The SILCS methodology allows for the generation of functional group affinity maps (FragMaps) of proteins that take into account contributions from protein desolvation, functional group desolvation, protein flexibility as well as direct interactions of the functional groups with the protein. Boltzmann transformation of the maps yields Grid Free Energy (GFE) FragMaps that may be used both qualitatively and quantitatively to direct ligand design. To allow for the application of the SILCS approach to deep and occluded pockets in proteins an oscillating μex Grand Canonical Monte Carlo (GCMC) approach was developed that allows for insertions of small solute molecules in the presence of an explicit aqueous environment. Combining the GCMC method with MD simulations for the inclusion of protein flexibility allows for the determination of GFE Fragmaps in occluded pockets. An overview of the GCMC/MD SILCS methodology along with application of the method to T4-lysozyme pocket mutants, nuclear receptors and GPCRs will be presented.
Alex MacKerellComputational functional group affinity mapping of proteins is of utility for ligand design in the context of database screening, fragment-based design and lead compound optimization. The SILCS methodology allows for the generation of functional group affinity maps (FragMaps) of proteins that take into account contributions from protein desolvation, functional group desolvation, protein flexibility as well as direct interactions of the functional groups with the protein. Boltzmann transformation of the maps yields Grid Free Energy (GFE) FragMaps that may be used both qualitatively and quantitatively to direct ligand design. To allow for the application of the SILCS approach to deep and occluded pockets in proteins an oscillating μex Grand Canonical Monte Carlo (GCMC) approach was developed that allows for insertions of small solute molecules in the presence of an explicit aqueous environment. Combining the GCMC method with MD simulations for the inclusion of protein flexibility allows for the determination of GFE Fragmaps in occluded pockets. An overview of the GCMC/MD SILCS methodology along with application of the method to T4-lysozyme pocket mutants, nuclear receptors and GPCRs will be presented. Sandor VajdaBinding energy hot spots, smaller regions of binding sites that contribute a disproportionate amount to the free energy of binding any ligand, can be determined computationally from ligand-free structures of protein targets. The hot spot structure of a protein target provides very useful information on binding properties. The first application that will be discussed is predicting druggability, i.e., the ability of a site of binding druglike ligands with sufficient affinity. We applied the method to a large set of proteins. Results showed that, because the method is based on the biophysics of binding rather than on empirical parameterization, meaningful information can be gained about classes of proteins and classes of compounds beyond those resembling validated targets and conventionally druglike ligands. In particular, the method identifies targets that, while not druggable by druglike compounds, may become druggable using compound classes such as macrocycles or other large molecules beyond the rule-of-five limit. Second, we show that the hot spots provide crucial insights into the prospects for successful application of fragment-based drug discovery (FBDD), and whether a fragment hit can be advanced into a high affinity ligand. The key factor is the strength of the top ranking hot spot, and how well a given fragment complements it. We show that published data are sufficient to provide a sophisticated and quantitative understanding of how hot spot strength, number and spatial arrangement govern the potential for a surface site to bind to fragment-sized and larger ligands. This improved understanding provides important guidance for the effective application of FBDD in drug discovery.
Sandor VajdaBinding energy hot spots, smaller regions of binding sites that contribute a disproportionate amount to the free energy of binding any ligand, can be determined computationally from ligand-free structures of protein targets. The hot spot structure of a protein target provides very useful information on binding properties. The first application that will be discussed is predicting druggability, i.e., the ability of a site of binding druglike ligands with sufficient affinity. We applied the method to a large set of proteins. Results showed that, because the method is based on the biophysics of binding rather than on empirical parameterization, meaningful information can be gained about classes of proteins and classes of compounds beyond those resembling validated targets and conventionally druglike ligands. In particular, the method identifies targets that, while not druggable by druglike compounds, may become druggable using compound classes such as macrocycles or other large molecules beyond the rule-of-five limit. Second, we show that the hot spots provide crucial insights into the prospects for successful application of fragment-based drug discovery (FBDD), and whether a fragment hit can be advanced into a high affinity ligand. The key factor is the strength of the top ranking hot spot, and how well a given fragment complements it. We show that published data are sufficient to provide a sophisticated and quantitative understanding of how hot spot strength, number and spatial arrangement govern the potential for a surface site to bind to fragment-sized and larger ligands. This improved understanding provides important guidance for the effective application of FBDD in drug discovery. Cameron AbramsPrediction of binding and unbinding kinetics from all-atom molecular simulations could one day be an important component in the toolkit for structure-based drug design. Unfortunately, in contrast to binding affinity prediction, algorithms for estimating on- and off-rates in relatively complicated systems remain relatively undeveloped. Here we demonstrate one approach that has been applied to successfully compute the entry and exit rates of small gas molecules into globular proteins. The method is an application of transition-path theory and involves performing milestoning MD simulations in cells forming a Voronoi tesselation of some chosen state-discriminating collective-variable space. Particular attention is paid to handshaking the simulations with a continuous approximation of the solute flux to a protein target surface as a function of ligand diffusivity and bulk concentration. The method is applicable beyond small gas molecules and extensions involving more complicated molecules will be discussed.
Cameron AbramsPrediction of binding and unbinding kinetics from all-atom molecular simulations could one day be an important component in the toolkit for structure-based drug design. Unfortunately, in contrast to binding affinity prediction, algorithms for estimating on- and off-rates in relatively complicated systems remain relatively undeveloped. Here we demonstrate one approach that has been applied to successfully compute the entry and exit rates of small gas molecules into globular proteins. The method is an application of transition-path theory and involves performing milestoning MD simulations in cells forming a Voronoi tesselation of some chosen state-discriminating collective-variable space. Particular attention is paid to handshaking the simulations with a continuous approximation of the solute flux to a protein target surface as a function of ligand diffusivity and bulk concentration. The method is applicable beyond small gas molecules and extensions involving more complicated molecules will be discussed. Chris ChipotOne of the grand challenges of rationale drug design is the prediction of the affinity of potential therapeutic agents for a given protein target. This challenge is in large measure rooted in the considerable changes in configurational entropy that accompanies the binding process, which atomistic simulations cannot easily sample. Two strategies relying upon alchemical transformations, on the one hand, and geometric transformations, notably potential of mean force calculations, on the other hand, are proposed, invoking a series of geometric restraints acting on collecting variables designed to alleviate sampling limitations inherent to classical molecular dynamics simulations. I will show through the example of a protein binding a small substrate, that both strategies, however of clearly different nature, can yield nearly identical standard binding free energies within chemical accuracy. I will further show how the methodology can be seamlessly transposed to protein-protein complexes. I will also outline current strategies to estimate binding entropies from such calculations. Downstream from the prediction of binding affinities is the challenging prediction of bioavailability. To estimate the permeability of the biological membrane to a drug candidate, an approach based upon Bayesian inferences, which reconciles thermodynamics and kinetics in molecular dynamics simulations with time-dependent biases, is put forth. Performance of the method is illustrated with prototypical permeants diffusing in a homogeneous lipid bilayer.
Chris ChipotOne of the grand challenges of rationale drug design is the prediction of the affinity of potential therapeutic agents for a given protein target. This challenge is in large measure rooted in the considerable changes in configurational entropy that accompanies the binding process, which atomistic simulations cannot easily sample. Two strategies relying upon alchemical transformations, on the one hand, and geometric transformations, notably potential of mean force calculations, on the other hand, are proposed, invoking a series of geometric restraints acting on collecting variables designed to alleviate sampling limitations inherent to classical molecular dynamics simulations. I will show through the example of a protein binding a small substrate, that both strategies, however of clearly different nature, can yield nearly identical standard binding free energies within chemical accuracy. I will further show how the methodology can be seamlessly transposed to protein-protein complexes. I will also outline current strategies to estimate binding entropies from such calculations. Downstream from the prediction of binding affinities is the challenging prediction of bioavailability. To estimate the permeability of the biological membrane to a drug candidate, an approach based upon Bayesian inferences, which reconciles thermodynamics and kinetics in molecular dynamics simulations with time-dependent biases, is put forth. Performance of the method is illustrated with prototypical permeants diffusing in a homogeneous lipid bilayer. Martin GoetheMinimizing a suitable free energy expression is arguably the most common approach in (ab initio) protein structure prediction. The achieved accuracy depends crucially on the quality of the free energy expression in use. Here, we present corrections to existing free energy expressions which arise from the thermal motion of the protein. We (i) devise a term accounting for the vibrational entropy of the protein, and (ii) correct existing potentials for the "thermal smoothing effect".
Martin GoetheMinimizing a suitable free energy expression is arguably the most common approach in (ab initio) protein structure prediction. The achieved accuracy depends crucially on the quality of the free energy expression in use. Here, we present corrections to existing free energy expressions which arise from the thermal motion of the protein. We (i) devise a term accounting for the vibrational entropy of the protein, and (ii) correct existing potentials for the "thermal smoothing effect". Tom KurtzmanDespite recent advances in methodologies that better characterize local water structure and thermodynamics, the simply- stated question of whether it would be beneficial or detrimental, in a free-energetic sense, to displace water from a region of a protein with a suitably complementary ligand continues to be a conundrum. Solvation thermodynamic mapping techniques have provided valuable insight into the role of displacing water in molecular recognition however solvation thermodynamics alone is not predictive of displaceability and tightly binding ligands regularly displace water from both regions that are characterized as having favorable solvation and regions that are characterized as having unfavorable solvation. Part of the difficulty of assessing the thermodynamics of water displacement is due to the large number of often counteracting contributions and the consequent lack of a simplifying conceptual framework. We will discuss the key insights which have been gained from solvation thermodynamic mapping and how we are incorporating them into drug discovery and design methodologies. We will also discuss remaining issues on predicting favorability of water displacement and suggest how they might be tackled.
Tom KurtzmanDespite recent advances in methodologies that better characterize local water structure and thermodynamics, the simply- stated question of whether it would be beneficial or detrimental, in a free-energetic sense, to displace water from a region of a protein with a suitably complementary ligand continues to be a conundrum. Solvation thermodynamic mapping techniques have provided valuable insight into the role of displacing water in molecular recognition however solvation thermodynamics alone is not predictive of displaceability and tightly binding ligands regularly displace water from both regions that are characterized as having favorable solvation and regions that are characterized as having unfavorable solvation. Part of the difficulty of assessing the thermodynamics of water displacement is due to the large number of often counteracting contributions and the consequent lack of a simplifying conceptual framework. We will discuss the key insights which have been gained from solvation thermodynamic mapping and how we are incorporating them into drug discovery and design methodologies. We will also discuss remaining issues on predicting favorability of water displacement and suggest how they might be tackled.