MBI Videos
Workshop 1: Dynamics in Networks with Special Properties
-
 Pauline van den DriesscheA network epidemic model for cholera and other diseases that can be transmitted via the environment is developed by adapting the Miller model to include the environment. The person-to-person contacts are modeled by a random contact network, and the contagious environment is modeled by an external node that connects to every individual. The dynamics of our model show excellent agreement with stochastic simulations. The basic reproduction number R0 is computed, and on a Poisson network shown to be the sum of the basic reproduction numbers of the person-to-person and person-to-water-to-person transmission pathways, as in the homogeneous mixing limit. How- ever, on other networks, R0 depends nonlinearly on the transmission along the two pathways. Type reproduction numbers are computed and quantify measures to control cholera.
Pauline van den DriesscheA network epidemic model for cholera and other diseases that can be transmitted via the environment is developed by adapting the Miller model to include the environment. The person-to-person contacts are modeled by a random contact network, and the contagious environment is modeled by an external node that connects to every individual. The dynamics of our model show excellent agreement with stochastic simulations. The basic reproduction number R0 is computed, and on a Poisson network shown to be the sum of the basic reproduction numbers of the person-to-person and person-to-water-to-person transmission pathways, as in the homogeneous mixing limit. How- ever, on other networks, R0 depends nonlinearly on the transmission along the two pathways. Type reproduction numbers are computed and quantify measures to control cholera. -
 Murad BanajiThe combinatorial structure of a chemical reaction network (CRN) may determine various behaviours of the associated dynamical systems. From network structure we may gain information about multistationarity, oscillation, bifurcations, persistence, distances between trajectories, orderings of variables, etc. A large number of questions naturally take the form of decision problems (formal languages): can a given network have some particular behaviour? Pursuing results in this area we are led to problems in graph theory, linear and exterior algebra, analysis, and geometry, sometimes of interest beyond their immediate application and intersecting work in other domains. Even where there are elegant characterisations of networks with some property, questions about the complexity of the associated decision problems often remain. I will outline some results and open problems in this area.
Murad BanajiThe combinatorial structure of a chemical reaction network (CRN) may determine various behaviours of the associated dynamical systems. From network structure we may gain information about multistationarity, oscillation, bifurcations, persistence, distances between trajectories, orderings of variables, etc. A large number of questions naturally take the form of decision problems (formal languages): can a given network have some particular behaviour? Pursuing results in this area we are led to problems in graph theory, linear and exterior algebra, analysis, and geometry, sometimes of interest beyond their immediate application and intersecting work in other domains. Even where there are elegant characterisations of networks with some property, questions about the complexity of the associated decision problems often remain. I will outline some results and open problems in this area. -
 Stephen CoombesThe master stability function is a powerful tool for determining synchrony in high dimensional networks of coupled limit cycle oscillators. In part this approach relies on the analysis of a low dimensional variational equation around a periodic orbit. For smooth dynamical systems this orbit is not generically available in closed form. However, many models in physics, engineering, and biology admit to piece-wise linear (pwl) caricatures which are also often nonsmooth, for which it is possible to construct periodic orbits without recourse to numerical evolution of trajectories. A classic example is the McKean model of an excitable system that has been extensively studied in the mathematical neuroscience community. Understandably the master stability function cannot be immediately applied to networks of such elements if they are non-smooth. Here we show how to extend the master stability function to nonsmooth planar pwl systems, and in the process demonstrate that considerable insight into network dynamics can be obtained when choosing the dynamics of the nodes to be pwl. In illustration we highlight an inverse period-doubling route to synchrony, under variation in coupling strength, in linearly coupled networks for which the node dynamics is poised near a homoclinic bifurcation. We contrast this with node dynamics poised near a non-smooth Andronov-Hopf bifurcation and also a saddle node bifurcation of limit cycles, for which no such bifurcation of synchrony occurs.
Stephen CoombesThe master stability function is a powerful tool for determining synchrony in high dimensional networks of coupled limit cycle oscillators. In part this approach relies on the analysis of a low dimensional variational equation around a periodic orbit. For smooth dynamical systems this orbit is not generically available in closed form. However, many models in physics, engineering, and biology admit to piece-wise linear (pwl) caricatures which are also often nonsmooth, for which it is possible to construct periodic orbits without recourse to numerical evolution of trajectories. A classic example is the McKean model of an excitable system that has been extensively studied in the mathematical neuroscience community. Understandably the master stability function cannot be immediately applied to networks of such elements if they are non-smooth. Here we show how to extend the master stability function to nonsmooth planar pwl systems, and in the process demonstrate that considerable insight into network dynamics can be obtained when choosing the dynamics of the nodes to be pwl. In illustration we highlight an inverse period-doubling route to synchrony, under variation in coupling strength, in linearly coupled networks for which the node dynamics is poised near a homoclinic bifurcation. We contrast this with node dynamics poised near a non-smooth Andronov-Hopf bifurcation and also a saddle node bifurcation of limit cycles, for which no such bifurcation of synchrony occurs. -
 Roderick EdwardsA theory for qualitative models of gene regulatory networks has been developed over several decades, generally considering transcription factors to regulate directly the expression of other transcription factors, without any intermediate variables. In fact, gene expression always involves transcription, which produces mRNA molecules, followed by translation, which produces protein molecules, and which can then act as transcription factors for other genes. Here we explore a class of models that explicitly includes both transcription and translation, keeping track of both mRNA and protein concentrations. We mainly deal with transcription regulation functions that are steep sigmoids or step functions, as is often done in protein-only models, though translation is governed by a linear term. We extend many aspects of the protein-only theory to this new context, including properties of fixed points, mappings between switching points, qualitative analysis via a state-transition diagram, and a result on periodic orbits for negative feedback loops. We find that while singular behaviour in switching domains is largely avoided, non-uniqueness of solutions can still occur in the step-function limit.
Roderick EdwardsA theory for qualitative models of gene regulatory networks has been developed over several decades, generally considering transcription factors to regulate directly the expression of other transcription factors, without any intermediate variables. In fact, gene expression always involves transcription, which produces mRNA molecules, followed by translation, which produces protein molecules, and which can then act as transcription factors for other genes. Here we explore a class of models that explicitly includes both transcription and translation, keeping track of both mRNA and protein concentrations. We mainly deal with transcription regulation functions that are steep sigmoids or step functions, as is often done in protein-only models, though translation is governed by a linear term. We extend many aspects of the protein-only theory to this new context, including properties of fixed points, mappings between switching points, qualitative analysis via a state-transition diagram, and a result on periodic orbits for negative feedback loops. We find that while singular behaviour in switching domains is largely avoided, non-uniqueness of solutions can still occur in the step-function limit. -
 Anne ShiuWhen taken with mass-action kinetics, which reaction networks admit multiple steady states? What structure must such a network possess? Mathematically, this question is: among certain parametrized families of polynomial systems, which families admit multiple positive roots (for some parameter values)? No complete answer is known, although various criteria now exist---some to answer the question in the affirmative and some in the negative. In this talk, we answer these questions for the smallest networks—those with only a few chemical species or reactions. Our results highlight the role played by the Newton polytope of a network (the convex hull of the reactant vectors). It has become apparent in recent years that analyzing this Newton polytope elucidates some aspects of the long-term dynamics and can be used to determine whether the network always admits at least one steady state. What is new here is our use of the geometric objects to determine whether a network admits steady state. Finally, our work is motivated by recent results that connect the capacity for multistationarity of a given network to that of certain related networks which are typically smaller: we are therefore interested in classifying small multistationary networks, and our results form the first step in this direction.
Anne ShiuWhen taken with mass-action kinetics, which reaction networks admit multiple steady states? What structure must such a network possess? Mathematically, this question is: among certain parametrized families of polynomial systems, which families admit multiple positive roots (for some parameter values)? No complete answer is known, although various criteria now exist---some to answer the question in the affirmative and some in the negative. In this talk, we answer these questions for the smallest networks—those with only a few chemical species or reactions. Our results highlight the role played by the Newton polytope of a network (the convex hull of the reactant vectors). It has become apparent in recent years that analyzing this Newton polytope elucidates some aspects of the long-term dynamics and can be used to determine whether the network always admits at least one steady state. What is new here is our use of the geometric objects to determine whether a network admits steady state. Finally, our work is motivated by recent results that connect the capacity for multistationarity of a given network to that of certain related networks which are typically smaller: we are therefore interested in classifying small multistationary networks, and our results form the first step in this direction. -
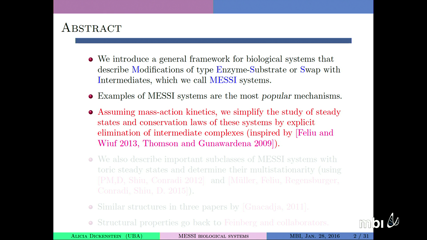 Alicia DickensteinWe introduce a general framework for biological systems that describe Modifications of type Enzyme-Substrate or Swap with Intermediates, which we call MESSI systems. Examples of MESSI systems are the sequential distributive or processive multisite phosphorylation networks, phosphorylation cascades, and the bacterial EnvZ/OmpR network. Assuming mass-action kinetics, we simplify the study of steady states and conservation laws of these systems by explicit elimination of intermediate complexes (inspired by [Feliu and Wiuf 2013, Thomson and Gunawardena 2009]). We also describe an important subclass of MESSI systems with toric steady states, for which we give combinatorial conditions to determine multistationarity and the occurrence of relevant boundary steady states. Joint work with Mercedes Pérez Millán.
Alicia DickensteinWe introduce a general framework for biological systems that describe Modifications of type Enzyme-Substrate or Swap with Intermediates, which we call MESSI systems. Examples of MESSI systems are the sequential distributive or processive multisite phosphorylation networks, phosphorylation cascades, and the bacterial EnvZ/OmpR network. Assuming mass-action kinetics, we simplify the study of steady states and conservation laws of these systems by explicit elimination of intermediate complexes (inspired by [Feliu and Wiuf 2013, Thomson and Gunawardena 2009]). We also describe an important subclass of MESSI systems with toric steady states, for which we give combinatorial conditions to determine multistationarity and the occurrence of relevant boundary steady states. Joint work with Mercedes Pérez Millán. -
 Manoj GopalkrishnanThe goal is to design an “intelligent chemical soup� that can do statistical inference. This may have niche technological applications in medicine and biological research, as well as provide fundamental insight into the workings of biochemical reaction pathways. As a first step towards our goal, we describe a scheme that exploits the remarkable mathematical similarity between log-linear models in statistics and chemical reaction networks. We present a simple scheme that encodes the information in a log-linear model as a chemical reaction network. Observed data is encoded as initial concentrations, and the equilibria of the corresponding mass-action system yield the maximum likelihood estimators. The simplicity of our scheme suggests that molecular environments, especially within cells, may be particularly well suited to performing statistical computations.
Manoj GopalkrishnanThe goal is to design an “intelligent chemical soup� that can do statistical inference. This may have niche technological applications in medicine and biological research, as well as provide fundamental insight into the workings of biochemical reaction pathways. As a first step towards our goal, we describe a scheme that exploits the remarkable mathematical similarity between log-linear models in statistics and chemical reaction networks. We present a simple scheme that encodes the information in a log-linear model as a chemical reaction network. Observed data is encoded as initial concentrations, and the equilibria of the corresponding mass-action system yield the maximum likelihood estimators. The simplicity of our scheme suggests that molecular environments, especially within cells, may be particularly well suited to performing statistical computations. -
![Andrey Shilnikov:Basic Bifurcation Theory for [small] Neural Networks - CPGs video photo](https://video.mbi.ohio-state.edu/vod/mbi-media/20160129-07/images/00000001.png) Andrey Shilnikov
Andrey Shilnikov -
 Patrick De LeenheerAbstract not submitted
Patrick De LeenheerAbstract not submitted -
 Fernando Antoneli
Fernando Antoneli -
 Stanislav ShvartsmanMultisite phosphorylation cycles are ubiquitous in cell regulation and are studied at multiple levels of complexity, with the ultimate goal to establish a quantitative view of phosphorylation networks in vivo. Achieving this goal is essentially impossible without mathematical models. Several models of multisite phosphorylation have been already proposed in the literature and received considerable attention from both experimentalists and theorists. Most of these models do not discriminate between distinct partially phosphorylated states of the regulated proteins and focus on two limiting regimes, distributive and processive, which differ in the number of enzyme substrate encounters needed for complete phosphorylation or dephosphorylation. Here we use the minimal model of ERK regulation to explore the dynamics of multisite phosphorylation in a reaction network that includes all essential phosphorylation states and varying levels of reaction processivity. In addition to bistability, which has been extensively studied in models with distributive mechanisms, this network can also generate oscillations, in which the relative abundances of the four phosphorylation states change in an ordered way. Both bistability and oscillations are suppressed at high levels of reaction processivity. Our work provides a general approach for large scale analysis of dynamics in multisite phosphorylation systems.
Stanislav ShvartsmanMultisite phosphorylation cycles are ubiquitous in cell regulation and are studied at multiple levels of complexity, with the ultimate goal to establish a quantitative view of phosphorylation networks in vivo. Achieving this goal is essentially impossible without mathematical models. Several models of multisite phosphorylation have been already proposed in the literature and received considerable attention from both experimentalists and theorists. Most of these models do not discriminate between distinct partially phosphorylated states of the regulated proteins and focus on two limiting regimes, distributive and processive, which differ in the number of enzyme substrate encounters needed for complete phosphorylation or dephosphorylation. Here we use the minimal model of ERK regulation to explore the dynamics of multisite phosphorylation in a reaction network that includes all essential phosphorylation states and varying levels of reaction processivity. In addition to bistability, which has been extensively studied in models with distributive mechanisms, this network can also generate oscillations, in which the relative abundances of the four phosphorylation states change in an ordered way. Both bistability and oscillations are suppressed at high levels of reaction processivity. Our work provides a general approach for large scale analysis of dynamics in multisite phosphorylation systems. -
 Matthias WolfrumSystems of coupled oscillators show a variety of collective dynamical regimes. We present a collection of such nonlinear phenomena, including non-universal transitions to synchrony in globally coupled oscillators, self-organized patterns of coherence and incoherence, called "chimera states" in spatially extended systems, and the emergence of macroscopic spatio-temporal chaos in such systems.
Matthias WolfrumSystems of coupled oscillators show a variety of collective dynamical regimes. We present a collection of such nonlinear phenomena, including non-universal transitions to synchrony in globally coupled oscillators, self-organized patterns of coherence and incoherence, called "chimera states" in spatially extended systems, and the emergence of macroscopic spatio-temporal chaos in such systems. -
 Konstantin MischaikowExperimental data on gene regulation is mostly qualitative, where the only information available about pairwise interactions is the presence of either up-or down- regulation. Quantitative data is often subject to large uncertainty and is mostly in terms of fold differences. Given these realities, it is very difficult to make reliable predictions using mathematical models. The current approach of choosing reasonable parameter values, a few initial conditions and then making predictions based on resulting solutions is severely subsampling both the parameter and phase space. This approach does not produce provable and reliable predictions.
Konstantin MischaikowExperimental data on gene regulation is mostly qualitative, where the only information available about pairwise interactions is the presence of either up-or down- regulation. Quantitative data is often subject to large uncertainty and is mostly in terms of fold differences. Given these realities, it is very difficult to make reliable predictions using mathematical models. The current approach of choosing reasonable parameter values, a few initial conditions and then making predictions based on resulting solutions is severely subsampling both the parameter and phase space. This approach does not produce provable and reliable predictions.
We present a new approach that uses continuous time Boolean networks as a platform for qualitative studies of gene regulation. In this talk we focus on the theoretical justification for the approach that we are taking. -
 Tomas GedeonExperimental data on gene regulation is mostly qualitative, where the only information available about pairwise interactions is the presence of either up-or down- regulation. Quantitative data is often subject to large uncertainty and is mostly in terms of fold differences. Given these realities, it is very difficult to make reliable predictions using mathematical models. The current approach of choosing reasonable parameter values, a few initial conditions and then making predictions based on resulting solutions is severely subsampling both the parameter and phase space. This approach does not produce provable and reliable predictions.
Tomas GedeonExperimental data on gene regulation is mostly qualitative, where the only information available about pairwise interactions is the presence of either up-or down- regulation. Quantitative data is often subject to large uncertainty and is mostly in terms of fold differences. Given these realities, it is very difficult to make reliable predictions using mathematical models. The current approach of choosing reasonable parameter values, a few initial conditions and then making predictions based on resulting solutions is severely subsampling both the parameter and phase space. This approach does not produce provable and reliable predictions.
We present a new approach that uses continuous time Boolean networks as a platform for qualitative studies of gene regulation. In this talk we show how we plan to use this approach in applications ranging from cell cycle dynamics to malaria. -
 Antonio PolitiAn ensemble of mean-field coupled oscillators characterized by different frequencies can exhibit a highly complex collective dynamics. I discuss an example where the phase-response curve is derived by smoothing out the response of delayed leaky integrate-and-fire neurons. It turns out that even though the microscopic dynamics is linearly stable, the global (macroscopic) evolution is irregular (high-dimensional). This poses the question of how the two levels of description are actually connected to one another.
Antonio PolitiAn ensemble of mean-field coupled oscillators characterized by different frequencies can exhibit a highly complex collective dynamics. I discuss an example where the phase-response curve is derived by smoothing out the response of delayed leaky integrate-and-fire neurons. It turns out that even though the microscopic dynamics is linearly stable, the global (macroscopic) evolution is irregular (high-dimensional). This poses the question of how the two levels of description are actually connected to one another. -
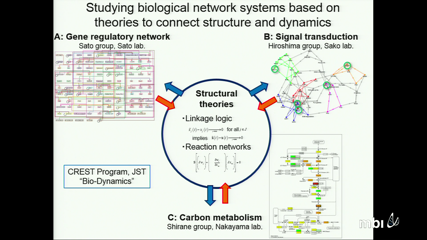 Atsushi MochizukiBy the success of modern biology we have many examples of large networks which describe interactions between a large number of species of bio-molecules. On the other hand, we have a limited understanding for quantitative details of biological systems, like the regulatory functions, parameter values of reaction rates. To overcome this problem, we have developed structural theories for dynamics of network systems. By our theories, important aspects of the dynamical properties of the system can be derived from information on the network structure, only, without assuming other quantitative details. In this talk, I will introduce a new theory for chemical reaction networks.
Atsushi MochizukiBy the success of modern biology we have many examples of large networks which describe interactions between a large number of species of bio-molecules. On the other hand, we have a limited understanding for quantitative details of biological systems, like the regulatory functions, parameter values of reaction rates. To overcome this problem, we have developed structural theories for dynamics of network systems. By our theories, important aspects of the dynamical properties of the system can be derived from information on the network structure, only, without assuming other quantitative details. In this talk, I will introduce a new theory for chemical reaction networks.
In living cells a large number of reactions are connected by sharing substrates or product chemicals, forming complex network systems like metabolic network. One experimental approach to the dynamics of such systems examines their sensitivity: each enzyme mediating a reaction in the system is increased/decreased or knocked out separately, and the responses in the concentrations of chemicals or their fluxes are observed. However, due to the complexity of the systems, it has been unclear how the network structures influence/determine the responses of the systems. In this study, we present a mathematical method, named structural sensitivity analysis, to determine the sensitivity of reaction systems from information on the network alone. We investigate how the sensitivity responses of chemicals in a reaction network depend on the structure of the network, and on the position of the perturbed reaction in the network. We establish and prove a general law which connects the network topology and the sensitivity patterns of metabolite responses directly. Our theorem explains two prominent features of network in sensitivity: localization and hierarchy in response pattern. We apply our method to several hypothetical and real life chemical reaction networks, including the metabolic network of the E. coli TCA cycle. The theorem is useful, practically, when examining real biological networks based on sensitivity experiments. -
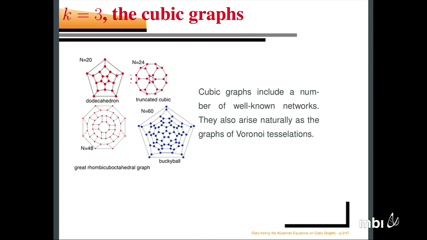 Bard ErmentroutIn this talk, I will describe the dynamics of a system of sinusoidally coupled phase oscillators on cubic graphs. The synchronous solution is always an attractor. However, as the graphs get larger (more nodes), it is possible to get other stable attractors. We study the basins, energy, and degree of stability of these non-synchronous attractors for all cubic graphs up to a certain number of nodes. We also use some techniques from computational algebraic geometry to show that for some graphs, the only attractor is synchrony.
Bard ErmentroutIn this talk, I will describe the dynamics of a system of sinusoidally coupled phase oscillators on cubic graphs. The synchronous solution is always an attractor. However, as the graphs get larger (more nodes), it is possible to get other stable attractors. We study the basins, energy, and degree of stability of these non-synchronous attractors for all cubic graphs up to a certain number of nodes. We also use some techniques from computational algebraic geometry to show that for some graphs, the only attractor is synchrony. -
 Maya MinchevaThe MAPK network is a principal component of many intracellular signaling modules. Multistability (the existence of multiple stable steady states) is considered an important property of such networks. Theoretical studies have established parameter values for multistability for many models of MAPK networks. Deciding if a given model has the capacity for multistationarity (the existence of multiple steady states) usually requires an extensive search of the parameter space. Two simple parameter inequalities will be presented. If the first inequality is satisfied, multistationarity, and hence the potential for multistability, is guaranteed. If the second inequality is satisfied, the uniqueness of a steady state, and hence the absence of multistability, is guaranteed. The method also allows for the direct computation of the total concentration values such that multistationarity occurs. Multistability in the ERK -- MEK -- MKP model will be presented. Some possible generalizations of this method will be discussed. This is a joint work with Carsten Conradi.
Maya MinchevaThe MAPK network is a principal component of many intracellular signaling modules. Multistability (the existence of multiple stable steady states) is considered an important property of such networks. Theoretical studies have established parameter values for multistability for many models of MAPK networks. Deciding if a given model has the capacity for multistationarity (the existence of multiple steady states) usually requires an extensive search of the parameter space. Two simple parameter inequalities will be presented. If the first inequality is satisfied, multistationarity, and hence the potential for multistability, is guaranteed. If the second inequality is satisfied, the uniqueness of a steady state, and hence the absence of multistability, is guaranteed. The method also allows for the direct computation of the total concentration values such that multistationarity occurs. Multistability in the ERK -- MEK -- MKP model will be presented. Some possible generalizations of this method will be discussed. This is a joint work with Carsten Conradi. -
 Pete AshwinThis talk will look at emergent "chimera" dynamics in coupled oscillator systems composed of identical and indistinguishable oscillators. We propose a checkable definition of a weak chimera state and give some basic results on systems that can/cannot have chimera states in their dynamics using this definition. These include chimera states for systems of at least four oscillators with two coupling strengths.
Pete AshwinThis talk will look at emergent "chimera" dynamics in coupled oscillator systems composed of identical and indistinguishable oscillators. We propose a checkable definition of a weak chimera state and give some basic results on systems that can/cannot have chimera states in their dynamics using this definition. These include chimera states for systems of at least four oscillators with two coupling strengths. -
 Jonathan RubinQuite a bit of work over many decades has gone into exploring respiratory rhythm-generation mechanisms. These studies have established an important role for the pre-Botzinger (pBC) complex in the mammalian brainstem and has investigated properties of single pBC neurons and their synaptic interactions. I will present work arising from efforts to understand how synchronous bursts of activity emerge across the network of pBC respiratory neurons. This work includes some computational approaches to systematically study how burst synchrony depends on network properties, some analytical approaches to estimate impacts of the prevalence of architectural motifs on the spread of activity in a network, and some rigorous analysis of graphicality and graph enumeration that are relevant to testing the motif-based ideas computationally.
Jonathan RubinQuite a bit of work over many decades has gone into exploring respiratory rhythm-generation mechanisms. These studies have established an important role for the pre-Botzinger (pBC) complex in the mammalian brainstem and has investigated properties of single pBC neurons and their synaptic interactions. I will present work arising from efforts to understand how synchronous bursts of activity emerge across the network of pBC respiratory neurons. This work includes some computational approaches to systematically study how burst synchrony depends on network properties, some analytical approaches to estimate impacts of the prevalence of architectural motifs on the spread of activity in a network, and some rigorous analysis of graphicality and graph enumeration that are relevant to testing the motif-based ideas computationally. -
 Alexey ZaikinI will discuss results of theoretical modelling in very multi-disciplinary area between Systems Medicine, Synthetic Biology, Artificial Intelligence and Applied Mathematics. Multicellular systems, e.g. neural networks of a living brain, can learn and be intelligent. Some of the principles of this intelligence have been mathematically formulated in the study of Artificial Intelligence (AI), starting from the basic Rosenblatt’s and associative Hebbian perceptrons and resulting in modern artificial neural networks with multilayer structure and recurrence. In some sense AI has mimicked the function of natural neural networks. However, relatively simple systems as cells are also able to perform tasks such as decision making and learning by utilizing their genetic regulatory frameworks. Intracellular genetic networks can be more intelligent than was first assumed due to their ability to learn. Such learning includes classification of several inputs or, the manifestations of this intelligence is the ability to learn associations of two stimuli within gene regulating circuitry: Hebbian type learning within the cellular life. However, gene expression is an intrinsically noisy process, hence, we investigate the effect of intrinsic and extrinsic noise on this kind of intracellular intelligence. During the talk I will also include brief introductions/tutorials about Synthetic Biology, modelling of genetic networks and noise-induced ordering.
Alexey ZaikinI will discuss results of theoretical modelling in very multi-disciplinary area between Systems Medicine, Synthetic Biology, Artificial Intelligence and Applied Mathematics. Multicellular systems, e.g. neural networks of a living brain, can learn and be intelligent. Some of the principles of this intelligence have been mathematically formulated in the study of Artificial Intelligence (AI), starting from the basic Rosenblatt’s and associative Hebbian perceptrons and resulting in modern artificial neural networks with multilayer structure and recurrence. In some sense AI has mimicked the function of natural neural networks. However, relatively simple systems as cells are also able to perform tasks such as decision making and learning by utilizing their genetic regulatory frameworks. Intracellular genetic networks can be more intelligent than was first assumed due to their ability to learn. Such learning includes classification of several inputs or, the manifestations of this intelligence is the ability to learn associations of two stimuli within gene regulating circuitry: Hebbian type learning within the cellular life. However, gene expression is an intrinsically noisy process, hence, we investigate the effect of intrinsic and extrinsic noise on this kind of intracellular intelligence. During the talk I will also include brief introductions/tutorials about Synthetic Biology, modelling of genetic networks and noise-induced ordering. -
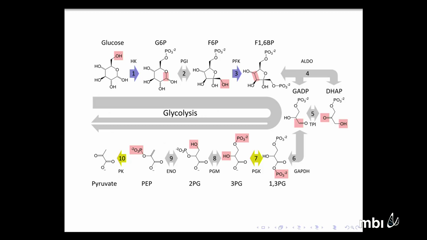 Badal JoshiIt is an open problem to identify reaction networks that admit multiple positive steady states. Criteria such as deficiency theory and Jacobian criterion help rule out the possibility of multiple steady states. But these tests are not sufficient to establish multistationarity. For fully open networks, we can establish multistationarity by relating the steady states of a reaction network with those of its component “embedded networks�. We refer to the multistationary fully open networks that are minimal with respect to the embedding relation as atoms of multistationarity. We identify some families of atoms of multistationarity and show that there exist arbitrarily large (in species, reactions) such atoms.
Badal JoshiIt is an open problem to identify reaction networks that admit multiple positive steady states. Criteria such as deficiency theory and Jacobian criterion help rule out the possibility of multiple steady states. But these tests are not sufficient to establish multistationarity. For fully open networks, we can establish multistationarity by relating the steady states of a reaction network with those of its component “embedded networks�. We refer to the multistationary fully open networks that are minimal with respect to the embedding relation as atoms of multistationarity. We identify some families of atoms of multistationarity and show that there exist arbitrarily large (in species, reactions) such atoms. -
 Marty Golubitsky
Marty GolubitskyNetworks of differential equations can be defined by directed graphs. The graphs (or network architecture) indicate who is talking to whom and when they are saying the same thing. We ask: Which properties of solutions of coupled equations follow from network architecture. Answers include "patterns of synchrony" for equilibria and "patterns of phase-shift synchrony" for time-periodic solutions. We show how these properties can be used to explain surprising results in binocular rivalry experiments and we discuss how homeostasis can be thought of as a network phenomenon.
-
 Yunjiao WangMultistable perception phenomena have been widely used for examining visual awareness and its underlying cortical mechanisms. Plausible models can explain binocular rivalry – the perceptual switching between two conflicting stimuli presented to each eye. Human subjects also report rivalry between percepts formed by grouping complementary patches from images presented to either eye. The dynamics of rivalry between such integrated percepts is not completely understood, and it is unclear whether models that explain binocular rivalry can be generalized. Classical models rely on mutual inhibition between distinct populations whose activity corresponds to each percept, with switches driven by adaptation or noise. Such models do not reflect the more complex patterns of neural activity necessary to describe interocular grouping. Moreover, the switching dynamics between more than two percepts is characterized by the sequence of perceptual states in addition to dominance times. Mechanistic models of multistable rivalry need to explain such dynamics.
Yunjiao WangMultistable perception phenomena have been widely used for examining visual awareness and its underlying cortical mechanisms. Plausible models can explain binocular rivalry – the perceptual switching between two conflicting stimuli presented to each eye. Human subjects also report rivalry between percepts formed by grouping complementary patches from images presented to either eye. The dynamics of rivalry between such integrated percepts is not completely understood, and it is unclear whether models that explain binocular rivalry can be generalized. Classical models rely on mutual inhibition between distinct populations whose activity corresponds to each percept, with switches driven by adaptation or noise. Such models do not reflect the more complex patterns of neural activity necessary to describe interocular grouping. Moreover, the switching dynamics between more than two percepts is characterized by the sequence of perceptual states in addition to dominance times. Mechanistic models of multistable rivalry need to explain such dynamics.
We studied the effect of color saturation on the dynamics of four-state perceptual rivalry. We presented subjects with split-grating stimuli composed of a half green grating and half red orthogonal grating to each eye. Subjects reliably reported four percepts: the two stimuli presented to each eye, as well as two coherent images resulting form interocular grouping. We hypothesized that an increase in color saturation would provide a strong cue to group the coherent halves, and would increase the dominance of grouped percepts. Experiments confirmed that this was the case. Further analysis showed that the increase in the fraction of time grouped stimuli were perceived was partly due to a decrease in single-eye dominance durations and partly due to an increase in the number of visits to grouped percepts. We used a computational model to show that our experimental observations can be reproduced by combining three mechanisms: mutual inhibition, recurrent excitation, and adaptation. -
 Stephen ProulxGene networks in living organisms are part of a dynamical system whose output make up the traits of organisms and determine reproductive fitness. One of the roles that such networks play is to respond to variability in the environment. Because organisms are the product of past evolution, we expect that evolution will generally increase organismal fitness, but this is subject to some constraints and historical effects. In this talk, I will discuss models of fluctuating environments where the output of the gene network determines fitness. I compare the outcome of optimal control models with evolved gene networks and discuss how the networks parameters evolve.
Stephen ProulxGene networks in living organisms are part of a dynamical system whose output make up the traits of organisms and determine reproductive fitness. One of the roles that such networks play is to respond to variability in the environment. Because organisms are the product of past evolution, we expect that evolution will generally increase organismal fitness, but this is subject to some constraints and historical effects. In this talk, I will discuss models of fluctuating environments where the output of the gene network determines fitness. I compare the outcome of optimal control models with evolved gene networks and discuss how the networks parameters evolve. -
 Georgi MedvedevIn this talk, I will discuss the continuum limit for coupled dynamical systems on large graphs and applications to stability of spatial patterns in the Kuramoto model of coupled phase oscillators.
Georgi MedvedevIn this talk, I will discuss the continuum limit for coupled dynamical systems on large graphs and applications to stability of spatial patterns in the Kuramoto model of coupled phase oscillators. -
 Matthew JohnstonSpurred by the rise of systems biology in the last decade and a half, network-based approaches have gained prominence as an efficient and insightful way to analyze complex biochemical reaction systems, such as MAPK signaling cascades and gene regulatory networks. Surprisingly, network-based methods are often able to make dynamical and steady state predictions independent of the initial conditions, rate parameters, and even rate form.
Matthew JohnstonSpurred by the rise of systems biology in the last decade and a half, network-based approaches have gained prominence as an efficient and insightful way to analyze complex biochemical reaction systems, such as MAPK signaling cascades and gene regulatory networks. Surprisingly, network-based methods are often able to make dynamical and steady state predictions independent of the initial conditions, rate parameters, and even rate form.
In this talk, I will outline some recent applications of generalized network theory to biochemical reaction systems. In a generalized network, there are two networks with the same topological structure: one for the stoichiometry, and one for the kinetics. Examples of biochemical reaction systems with dynamically equivalent but better structured generalized networks will be presented. -
 Casian PanteaThe capacity of reaction network system to exhibit two or more steady states has been the focus of considerable recent work. The question of multistationarity is closely related to that of injectivity of the corresponding vector field. In this talk we give an overview of some old and new results on injectivity and multistationarity in vector fields associated with interaction networks, under more or less general assumptions on the nature of the network and the kinetic laws. This is joint work with Murad Banaji.
Casian PanteaThe capacity of reaction network system to exhibit two or more steady states has been the focus of considerable recent work. The question of multistationarity is closely related to that of injectivity of the corresponding vector field. In this talk we give an overview of some old and new results on injectivity and multistationarity in vector fields associated with interaction networks, under more or less general assumptions on the nature of the network and the kinetic laws. This is joint work with Murad Banaji.
