-
Sayan Mukherjee
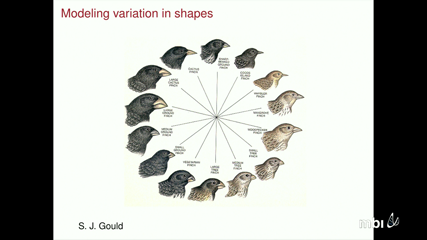
I will introduce a geometric/topological transform that allows for the modeling of shapes and surfaces without requiring landmarks. We discuss applications of this to morphological analysis of primates as well as the analysis of melanoma. I will also outline outline how the transform can measure distances between shapes as well as place probability models on shapes and surfaces. I will also discuss approaches using conformal geometry to model surfaces.
-
Washington Mio
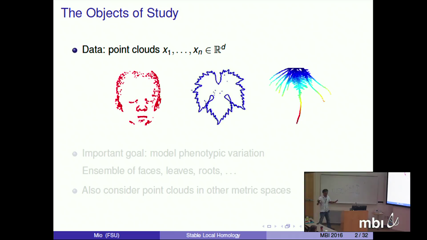
Persistent homology is a powerful technique for probing and analyzing the shape of data with complex distributions and has been widely used in studies of global organization of data across spatial scales. Much richer information can be uncovered through local and regional topology; however, naive localization is prone to instabilities as localization can be extremely sensitive to sampling and noise. In this talk, I will present an approach to local homology that is provably robust and discuss how it may be used in shape analysis, particularly in situations where invariance under group actions needs to be taken into account. I will begin with the case of Euclidean data and then discuss an extension to data on other metric spaces. In this formulation, local homology across scales is viewed as a path of barcodes or persistence diagrams that is stable with respect to the Wasserstein distance. In addition to illustrations using synthetic data, I will present an application to quantitative trait loci analysis of tomato leaf shape, a collaboration with researchers at the Danforth Plant Research Center. Here, the primary goal is to discover interpretable associations between genotypes and complex phenotypes to elucidate the genetic basis of plant morphology.
-

Steve Haase
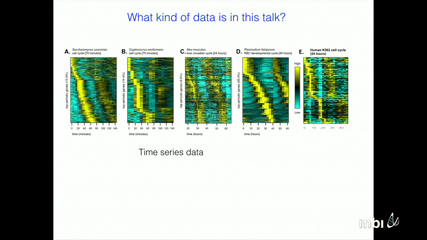
Gene regulatory networks (GRNs) can drive cyclic and temporally ordered processes in biological systems. One of the best-known GRNs of this type is the circadian clock, which drives rhythmic behaviors with a period of approximately 24hrs. The circadian clock network exerts its control, in part, by regulating a dynamic program of gene expression where substantial fractions of the genome are expressed during distinct phases of the circadian cycle. More recently, we have proposed that a different GRN controls the cyclic program of gene expression that is observed during the cell division cycle. In fact, we have observed similar gene expression programs across time scales from hours to days, and across organisms that are evolutionarily diverged by millions of years. These observations suggest that a class of GRNs may serve as central mechanisms that drive temporal gene expression programs in biological systems. One goal of our work is to identify the structure and function of these networks. New approaches for inferring the structure of these GRNs directly from time-series transcriptome data will be discussed. We will also describe experimental and quantitative approaches aimed at probing the dynamics of a GRN that controls the well-ordered, periodic program of transcription observed during the yeast cell-division cycle.
-

Facundo Memoli
We study methods for computing two network features with topological underpinnings: the Rips and Dowker Persistent Homology Diagrams. Our formulations work for general networks, which may be asymmetric and may have any real number as an edge weight. We study the sensitivity of Dowker persistence diagrams to intrinsic asymmetry in the data, and investigate the theoretical stability properties of both the Dowker and Rips persistence diagrams. We show experimental results on a variety of simulated and real world datasets using our methods. In particular, we apply both methods to a classification task on a database of networks.
-

Tim Sauer
Persistent homology has become a standard tool in modern data analysis, identifying homology generators at various scales as a parameter representing distances between points is increased. We show that for general sampling from a Riemannian manifold, there is a graph construction that captures all topological features in a single graph, which we call `consistent' homology. More precisely, the graph converges spectrally to the Laplace-De Rham operator of the manifold in the limit of large data. The graph construction, called continuous k-nearest neighbors (CkNN), neutralizes nonuniform sampling, and in practice reduces data requirements as well. We examine under which circumstances persistent or consistent approaches are preferred, and illustrate with data from neural cultures and physical experiments.
-

Chad Giusti
-

Vladimir Itskov
A convex code is a subset of the power set 2^n that arises from intersection patterns of convex sets in a Euclidean space. Despite their natural definition, these codes have not been studied until very recently, motivated by applications in neuroscience. Many neural systems generate patterns of neural activity that can be characterized mathematically as convex codes. These codes reflect topological features of the underlying stimulus space, some of which can be inferred using existing TDA methods. Perhaps surprisingly, not all codes can be realized by a convex cover, and the problem of determining which codes are convex is still open. I will begin by reviewing recent results on convex codes and the special case of hyperplane codes. I will then explain how hyperplane codes are related to the problem of detecting a feedforward network factorization using Dowker complexes, and present some computational results on this problem. Finally, I will describe an approach for detecting convexity of noisy neural codes that can be implemented using existing TDA algorithms.
-

Christopher Hillar
Discrete recurrent neural networks (DRNNs) were born in 1943 with the publication of the now seminal work by McCulloch and Pitts: "A logical calculus of the ideas immanent in nervous activity". Although the concepts led to major applications (digital circuit design, finite automata, computational theories of mind, Hopfield networks), experimental neuroscience has yet to benefit significantly. Here, we describe a novel, scalable use of DRNNs for the unsupervised discovery of structure in high-dimensional recordings of nervous tissue. We also present two case-studies in detail using the technology: (1) clustering of reoccurring spatiotemporal patterns in spike trains, and (2) denoising microscopy recordings of slices of neural activity. We also explain how to perform these analyses on standard hardware using our open-source Python package HDNET, which provides efficient DRNN tools for experimental neuroscientists. (Joint work with F. Effenberger).
-

Christine Heitsch
-
Guowei Wei
Biology is believed to be the last forefront of natural sciences. Recent advances in biotechnologies have led to the exponential growth of biological data, which paves the way for biological sciences to transform from qualitative, phenomenological and descriptive to quantitative, analytical and predictive. Mathematics is becoming a driven force behind this historic transformation as it did for quantum physics a century ago. I will discuss how to combine differential geometry, algebraic topology, graph theory and partial differential equation with machine learning to give rise to the most accurate predictions of tens of thousands of experimental data in solvation free energy, partition coefficient, protein-drug binding affinity, and protein mutation impact.
-
Tomas Gedeon
Experimental data on gene regulation is mostly qualitative, where the only information available about pairwise interactions is the presence of either up-or down- regulation. Quantitative data is often subject to large uncertainty and is mostly in terms of fold differences. Given these realities, it is very difficult to make reliable predictions using mathematical models. The current approach of choosing reasonable parameter values, a few initial conditions and then making predictions based on resulting solutions is severely subsampling both the parameter and phase space. This approach does not produce provable and reliable predictions.
We present a new approach that uses continuous time Boolean networks as a platform for qualitative studies of gene regulation. We compute a Database for Dynamics, which rigorously approximates global dynamics over entire parameter space. The results obtained by this method provably capture the dynamics at a predetermined spatial scale.
We apply our approach to study neighborhood of a given network in the space of networks. We start with a E2F-Rb network underlying the mammalian cell cycle restriction point and show that majority of the parameters support either the GO, NO-GO, or bistability between these two states. We then sample 100 perturbations of this network and study robustness of this dynamics in the network space.
-

Jessi Cisewski
Complicated spatial structures (CSS) are common in biological data (e.g. fibrin clots, fibroblasts), but are difficult to quantitatively analyze without losing important information. Topological data analysis (TDA) provides a way for biologists to better understand, visualize, and interpret such data. TDA is a statistical framework for extracting topological information from data and using it to estimate properties of the underlying structures. It has potential to dramatically improve the analysis of biological data by retrieving and quantifying crucial information that is missed in ad-hoc methods by specifically targeting shape-related features.
We present a framework for hypothesis testing of CSS using persistent homology. The randomness in the data (due to measurement error or topological noise) is transferred to randomness in the topological summaries, which provides an infrastructure for inference. These tests allow for statistical comparisons between CSS. We present several possible test statistics using persistence diagrams and carryout a simulation study to investigate the suitableness of the proposed test statistics.
-

Peter Bubenik
One approach to combining geometry, topology and statistics in the analysis of data consists of the following steps: (1) use the data to construct a geometric object; (2) apply topology to obtain a summary; and (3) apply statistics to the resulting summaries. From a statistical viewpoint, it is fruitful to replace the standard topological summary, the persistence diagram, with a vector (or better yet, a point in a Hilbert space). One such construction with particularly nice properties (e.g. reversability) is the persistence landscape. I will give an overview of this pipeline and apply it to analyze protein data and brain imaging data.
-
Ezra Miller
Biological data, such as the images of fruit fly wing veins that drive the ongoing investigations reported in this talk, generate persistent homology with multiple parameters each of which varies continuously. Statistical analysis of persistence in this context presents fundamental challenges, such as how to encode persistence summaries for automatic computation and how to carry out statistical analyses with the summaries---theoretically and algorithmically---particularly in view of nontrivial moduli for multiparameter persistence diagrams. This talk presents an algebraic and geometric framework that renders these challenges surmountable while also clarifying the topological interpretation of each multiparameter persistence summary. The framework is new and useful already for two discrete parameters but works equally well for continuous parameters, or even for filtrations by arbitrary partially ordered sets. Joint work with David Houle (Biology, Florida State), Ashleigh Thomas (grad student, Duke Math), and Justin Curry (postdoc, Duke Math).
-
J. S. Marron
Object Oriented Data Analysis is the statistical analysis of populations of complex objects. In the special case of Functional Data Analysis, these data objects are curves, where standard Euclidean approaches, such as principal components analysis, have been very successful. Challenges in modern medical image analysis motivate the statistical analysis of populations of more complex data objects which are elements of mildly non-Euclidean spaces, such as Lie Groups and Symmetric Spaces, or of strongly non-Euclidean spaces, such as spaces of tree-structured data objects. These new contexts for Object Oriented Data Analysis create several potentially large new interfaces between mathematics and statistics. The notion of Object Oriented Data Analysis also impacts data analysis, through providing a language for discussion of the many choices needed in many modern complex data analyses.
-

Sebastian Kurtek
We present a new Riemannian framework for comprehensive statistical shape analysis of 3D objects, represented by their boundaries (parameterized surfaces). By comprehensive framework, we mean tools for registration, comparison, averaging, and modeling of observed surfaces. Registration is analogous to removing all shape preserving transformations, which include translation, scale, rotation and re-parameterization. This framework is based on a special representation of surfaces termed square-root normal fields and a closely related elastic metric. The main advantages of this method are: (1) the elastic metric provides a natural interpretation of shape deformations that are being quantified, (2) this metric is invariant to re-parameterizations of surfaces, and (3) under the square-root normal field transformation, the complicated elastic metric becomes the standard L2 metric, simplifying parts of the implementation. We present numerous examples of shape comparisons for various types of surfaces in different application areas. We also compute average shapes, covariances and perform principal component analysis to explore the variability in different shape classes. These quantities are used to define generative shape models and for random sampling. Specifically, we showcase the applicability of the proposed framework in shape analysis of anatomical structures in different medical applications including Attention Deficit Hyperactivity Disorder and endometriosis.
-

Wojciech Chacholski
I will describe a method of constructing continuous invariants of multidimensional persistence modules using so called noise systems. My aim for this talk is to discuss questions related to stability and computability of such invariants.
 Sayan MukherjeeI will introduce a geometric/topological transform that allows for the modeling of shapes and surfaces without requiring landmarks. We discuss applications of this to morphological analysis of primates as well as the analysis of melanoma. I will also outline outline how the transform can measure distances between shapes as well as place probability models on shapes and surfaces. I will also discuss approaches using conformal geometry to model surfaces.
Sayan MukherjeeI will introduce a geometric/topological transform that allows for the modeling of shapes and surfaces without requiring landmarks. We discuss applications of this to morphological analysis of primates as well as the analysis of melanoma. I will also outline outline how the transform can measure distances between shapes as well as place probability models on shapes and surfaces. I will also discuss approaches using conformal geometry to model surfaces. Washington MioPersistent homology is a powerful technique for probing and analyzing the shape of data with complex distributions and has been widely used in studies of global organization of data across spatial scales. Much richer information can be uncovered through local and regional topology; however, naive localization is prone to instabilities as localization can be extremely sensitive to sampling and noise. In this talk, I will present an approach to local homology that is provably robust and discuss how it may be used in shape analysis, particularly in situations where invariance under group actions needs to be taken into account. I will begin with the case of Euclidean data and then discuss an extension to data on other metric spaces. In this formulation, local homology across scales is viewed as a path of barcodes or persistence diagrams that is stable with respect to the Wasserstein distance. In addition to illustrations using synthetic data, I will present an application to quantitative trait loci analysis of tomato leaf shape, a collaboration with researchers at the Danforth Plant Research Center. Here, the primary goal is to discover interpretable associations between genotypes and complex phenotypes to elucidate the genetic basis of plant morphology.
Washington MioPersistent homology is a powerful technique for probing and analyzing the shape of data with complex distributions and has been widely used in studies of global organization of data across spatial scales. Much richer information can be uncovered through local and regional topology; however, naive localization is prone to instabilities as localization can be extremely sensitive to sampling and noise. In this talk, I will present an approach to local homology that is provably robust and discuss how it may be used in shape analysis, particularly in situations where invariance under group actions needs to be taken into account. I will begin with the case of Euclidean data and then discuss an extension to data on other metric spaces. In this formulation, local homology across scales is viewed as a path of barcodes or persistence diagrams that is stable with respect to the Wasserstein distance. In addition to illustrations using synthetic data, I will present an application to quantitative trait loci analysis of tomato leaf shape, a collaboration with researchers at the Danforth Plant Research Center. Here, the primary goal is to discover interpretable associations between genotypes and complex phenotypes to elucidate the genetic basis of plant morphology. Steve HaaseGene regulatory networks (GRNs) can drive cyclic and temporally ordered processes in biological systems. One of the best-known GRNs of this type is the circadian clock, which drives rhythmic behaviors with a period of approximately 24hrs. The circadian clock network exerts its control, in part, by regulating a dynamic program of gene expression where substantial fractions of the genome are expressed during distinct phases of the circadian cycle. More recently, we have proposed that a different GRN controls the cyclic program of gene expression that is observed during the cell division cycle. In fact, we have observed similar gene expression programs across time scales from hours to days, and across organisms that are evolutionarily diverged by millions of years. These observations suggest that a class of GRNs may serve as central mechanisms that drive temporal gene expression programs in biological systems. One goal of our work is to identify the structure and function of these networks. New approaches for inferring the structure of these GRNs directly from time-series transcriptome data will be discussed. We will also describe experimental and quantitative approaches aimed at probing the dynamics of a GRN that controls the well-ordered, periodic program of transcription observed during the yeast cell-division cycle.
Steve HaaseGene regulatory networks (GRNs) can drive cyclic and temporally ordered processes in biological systems. One of the best-known GRNs of this type is the circadian clock, which drives rhythmic behaviors with a period of approximately 24hrs. The circadian clock network exerts its control, in part, by regulating a dynamic program of gene expression where substantial fractions of the genome are expressed during distinct phases of the circadian cycle. More recently, we have proposed that a different GRN controls the cyclic program of gene expression that is observed during the cell division cycle. In fact, we have observed similar gene expression programs across time scales from hours to days, and across organisms that are evolutionarily diverged by millions of years. These observations suggest that a class of GRNs may serve as central mechanisms that drive temporal gene expression programs in biological systems. One goal of our work is to identify the structure and function of these networks. New approaches for inferring the structure of these GRNs directly from time-series transcriptome data will be discussed. We will also describe experimental and quantitative approaches aimed at probing the dynamics of a GRN that controls the well-ordered, periodic program of transcription observed during the yeast cell-division cycle. Facundo MemoliWe study methods for computing two network features with topological underpinnings: the Rips and Dowker Persistent Homology Diagrams. Our formulations work for general networks, which may be asymmetric and may have any real number as an edge weight. We study the sensitivity of Dowker persistence diagrams to intrinsic asymmetry in the data, and investigate the theoretical stability properties of both the Dowker and Rips persistence diagrams. We show experimental results on a variety of simulated and real world datasets using our methods. In particular, we apply both methods to a classification task on a database of networks.
Facundo MemoliWe study methods for computing two network features with topological underpinnings: the Rips and Dowker Persistent Homology Diagrams. Our formulations work for general networks, which may be asymmetric and may have any real number as an edge weight. We study the sensitivity of Dowker persistence diagrams to intrinsic asymmetry in the data, and investigate the theoretical stability properties of both the Dowker and Rips persistence diagrams. We show experimental results on a variety of simulated and real world datasets using our methods. In particular, we apply both methods to a classification task on a database of networks. Tim SauerPersistent homology has become a standard tool in modern data analysis, identifying homology generators at various scales as a parameter representing distances between points is increased. We show that for general sampling from a Riemannian manifold, there is a graph construction that captures all topological features in a single graph, which we call `consistent' homology. More precisely, the graph converges spectrally to the Laplace-De Rham operator of the manifold in the limit of large data. The graph construction, called continuous k-nearest neighbors (CkNN), neutralizes nonuniform sampling, and in practice reduces data requirements as well. We examine under which circumstances persistent or consistent approaches are preferred, and illustrate with data from neural cultures and physical experiments.
Tim SauerPersistent homology has become a standard tool in modern data analysis, identifying homology generators at various scales as a parameter representing distances between points is increased. We show that for general sampling from a Riemannian manifold, there is a graph construction that captures all topological features in a single graph, which we call `consistent' homology. More precisely, the graph converges spectrally to the Laplace-De Rham operator of the manifold in the limit of large data. The graph construction, called continuous k-nearest neighbors (CkNN), neutralizes nonuniform sampling, and in practice reduces data requirements as well. We examine under which circumstances persistent or consistent approaches are preferred, and illustrate with data from neural cultures and physical experiments. Chad Giusti
Chad Giusti Vladimir ItskovA convex code is a subset of the power set 2^n that arises from intersection patterns of convex sets in a Euclidean space. Despite their natural definition, these codes have not been studied until very recently, motivated by applications in neuroscience. Many neural systems generate patterns of neural activity that can be characterized mathematically as convex codes. These codes reflect topological features of the underlying stimulus space, some of which can be inferred using existing TDA methods. Perhaps surprisingly, not all codes can be realized by a convex cover, and the problem of determining which codes are convex is still open. I will begin by reviewing recent results on convex codes and the special case of hyperplane codes. I will then explain how hyperplane codes are related to the problem of detecting a feedforward network factorization using Dowker complexes, and present some computational results on this problem. Finally, I will describe an approach for detecting convexity of noisy neural codes that can be implemented using existing TDA algorithms.
Vladimir ItskovA convex code is a subset of the power set 2^n that arises from intersection patterns of convex sets in a Euclidean space. Despite their natural definition, these codes have not been studied until very recently, motivated by applications in neuroscience. Many neural systems generate patterns of neural activity that can be characterized mathematically as convex codes. These codes reflect topological features of the underlying stimulus space, some of which can be inferred using existing TDA methods. Perhaps surprisingly, not all codes can be realized by a convex cover, and the problem of determining which codes are convex is still open. I will begin by reviewing recent results on convex codes and the special case of hyperplane codes. I will then explain how hyperplane codes are related to the problem of detecting a feedforward network factorization using Dowker complexes, and present some computational results on this problem. Finally, I will describe an approach for detecting convexity of noisy neural codes that can be implemented using existing TDA algorithms. Christopher HillarDiscrete recurrent neural networks (DRNNs) were born in 1943 with the publication of the now seminal work by McCulloch and Pitts: "A logical calculus of the ideas immanent in nervous activity". Although the concepts led to major applications (digital circuit design, finite automata, computational theories of mind, Hopfield networks), experimental neuroscience has yet to benefit significantly. Here, we describe a novel, scalable use of DRNNs for the unsupervised discovery of structure in high-dimensional recordings of nervous tissue. We also present two case-studies in detail using the technology: (1) clustering of reoccurring spatiotemporal patterns in spike trains, and (2) denoising microscopy recordings of slices of neural activity. We also explain how to perform these analyses on standard hardware using our open-source Python package HDNET, which provides efficient DRNN tools for experimental neuroscientists. (Joint work with F. Effenberger).
Christopher HillarDiscrete recurrent neural networks (DRNNs) were born in 1943 with the publication of the now seminal work by McCulloch and Pitts: "A logical calculus of the ideas immanent in nervous activity". Although the concepts led to major applications (digital circuit design, finite automata, computational theories of mind, Hopfield networks), experimental neuroscience has yet to benefit significantly. Here, we describe a novel, scalable use of DRNNs for the unsupervised discovery of structure in high-dimensional recordings of nervous tissue. We also present two case-studies in detail using the technology: (1) clustering of reoccurring spatiotemporal patterns in spike trains, and (2) denoising microscopy recordings of slices of neural activity. We also explain how to perform these analyses on standard hardware using our open-source Python package HDNET, which provides efficient DRNN tools for experimental neuroscientists. (Joint work with F. Effenberger). Christine Heitsch
Christine Heitsch Guowei WeiBiology is believed to be the last forefront of natural sciences. Recent advances in biotechnologies have led to the exponential growth of biological data, which paves the way for biological sciences to transform from qualitative, phenomenological and descriptive to quantitative, analytical and predictive. Mathematics is becoming a driven force behind this historic transformation as it did for quantum physics a century ago. I will discuss how to combine differential geometry, algebraic topology, graph theory and partial differential equation with machine learning to give rise to the most accurate predictions of tens of thousands of experimental data in solvation free energy, partition coefficient, protein-drug binding affinity, and protein mutation impact.
Guowei WeiBiology is believed to be the last forefront of natural sciences. Recent advances in biotechnologies have led to the exponential growth of biological data, which paves the way for biological sciences to transform from qualitative, phenomenological and descriptive to quantitative, analytical and predictive. Mathematics is becoming a driven force behind this historic transformation as it did for quantum physics a century ago. I will discuss how to combine differential geometry, algebraic topology, graph theory and partial differential equation with machine learning to give rise to the most accurate predictions of tens of thousands of experimental data in solvation free energy, partition coefficient, protein-drug binding affinity, and protein mutation impact. Tomas GedeonExperimental data on gene regulation is mostly qualitative, where the only information available about pairwise interactions is the presence of either up-or down- regulation. Quantitative data is often subject to large uncertainty and is mostly in terms of fold differences. Given these realities, it is very difficult to make reliable predictions using mathematical models. The current approach of choosing reasonable parameter values, a few initial conditions and then making predictions based on resulting solutions is severely subsampling both the parameter and phase space. This approach does not produce provable and reliable predictions.
Tomas GedeonExperimental data on gene regulation is mostly qualitative, where the only information available about pairwise interactions is the presence of either up-or down- regulation. Quantitative data is often subject to large uncertainty and is mostly in terms of fold differences. Given these realities, it is very difficult to make reliable predictions using mathematical models. The current approach of choosing reasonable parameter values, a few initial conditions and then making predictions based on resulting solutions is severely subsampling both the parameter and phase space. This approach does not produce provable and reliable predictions. Jessi CisewskiComplicated spatial structures (CSS) are common in biological data (e.g. fibrin clots, fibroblasts), but are difficult to quantitatively analyze without losing important information. Topological data analysis (TDA) provides a way for biologists to better understand, visualize, and interpret such data. TDA is a statistical framework for extracting topological information from data and using it to estimate properties of the underlying structures. It has potential to dramatically improve the analysis of biological data by retrieving and quantifying crucial information that is missed in ad-hoc methods by specifically targeting shape-related features.
Jessi CisewskiComplicated spatial structures (CSS) are common in biological data (e.g. fibrin clots, fibroblasts), but are difficult to quantitatively analyze without losing important information. Topological data analysis (TDA) provides a way for biologists to better understand, visualize, and interpret such data. TDA is a statistical framework for extracting topological information from data and using it to estimate properties of the underlying structures. It has potential to dramatically improve the analysis of biological data by retrieving and quantifying crucial information that is missed in ad-hoc methods by specifically targeting shape-related features. Peter BubenikOne approach to combining geometry, topology and statistics in the analysis of data consists of the following steps: (1) use the data to construct a geometric object; (2) apply topology to obtain a summary; and (3) apply statistics to the resulting summaries. From a statistical viewpoint, it is fruitful to replace the standard topological summary, the persistence diagram, with a vector (or better yet, a point in a Hilbert space). One such construction with particularly nice properties (e.g. reversability) is the persistence landscape. I will give an overview of this pipeline and apply it to analyze protein data and brain imaging data.
Peter BubenikOne approach to combining geometry, topology and statistics in the analysis of data consists of the following steps: (1) use the data to construct a geometric object; (2) apply topology to obtain a summary; and (3) apply statistics to the resulting summaries. From a statistical viewpoint, it is fruitful to replace the standard topological summary, the persistence diagram, with a vector (or better yet, a point in a Hilbert space). One such construction with particularly nice properties (e.g. reversability) is the persistence landscape. I will give an overview of this pipeline and apply it to analyze protein data and brain imaging data. Ezra MillerBiological data, such as the images of fruit fly wing veins that drive the ongoing investigations reported in this talk, generate persistent homology with multiple parameters each of which varies continuously. Statistical analysis of persistence in this context presents fundamental challenges, such as how to encode persistence summaries for automatic computation and how to carry out statistical analyses with the summaries---theoretically and algorithmically---particularly in view of nontrivial moduli for multiparameter persistence diagrams. This talk presents an algebraic and geometric framework that renders these challenges surmountable while also clarifying the topological interpretation of each multiparameter persistence summary. The framework is new and useful already for two discrete parameters but works equally well for continuous parameters, or even for filtrations by arbitrary partially ordered sets. Joint work with David Houle (Biology, Florida State), Ashleigh Thomas (grad student, Duke Math), and Justin Curry (postdoc, Duke Math).
Ezra MillerBiological data, such as the images of fruit fly wing veins that drive the ongoing investigations reported in this talk, generate persistent homology with multiple parameters each of which varies continuously. Statistical analysis of persistence in this context presents fundamental challenges, such as how to encode persistence summaries for automatic computation and how to carry out statistical analyses with the summaries---theoretically and algorithmically---particularly in view of nontrivial moduli for multiparameter persistence diagrams. This talk presents an algebraic and geometric framework that renders these challenges surmountable while also clarifying the topological interpretation of each multiparameter persistence summary. The framework is new and useful already for two discrete parameters but works equally well for continuous parameters, or even for filtrations by arbitrary partially ordered sets. Joint work with David Houle (Biology, Florida State), Ashleigh Thomas (grad student, Duke Math), and Justin Curry (postdoc, Duke Math). J. S. MarronObject Oriented Data Analysis is the statistical analysis of populations of complex objects. In the special case of Functional Data Analysis, these data objects are curves, where standard Euclidean approaches, such as principal components analysis, have been very successful. Challenges in modern medical image analysis motivate the statistical analysis of populations of more complex data objects which are elements of mildly non-Euclidean spaces, such as Lie Groups and Symmetric Spaces, or of strongly non-Euclidean spaces, such as spaces of tree-structured data objects. These new contexts for Object Oriented Data Analysis create several potentially large new interfaces between mathematics and statistics. The notion of Object Oriented Data Analysis also impacts data analysis, through providing a language for discussion of the many choices needed in many modern complex data analyses.
J. S. MarronObject Oriented Data Analysis is the statistical analysis of populations of complex objects. In the special case of Functional Data Analysis, these data objects are curves, where standard Euclidean approaches, such as principal components analysis, have been very successful. Challenges in modern medical image analysis motivate the statistical analysis of populations of more complex data objects which are elements of mildly non-Euclidean spaces, such as Lie Groups and Symmetric Spaces, or of strongly non-Euclidean spaces, such as spaces of tree-structured data objects. These new contexts for Object Oriented Data Analysis create several potentially large new interfaces between mathematics and statistics. The notion of Object Oriented Data Analysis also impacts data analysis, through providing a language for discussion of the many choices needed in many modern complex data analyses. Sebastian KurtekWe present a new Riemannian framework for comprehensive statistical shape analysis of 3D objects, represented by their boundaries (parameterized surfaces). By comprehensive framework, we mean tools for registration, comparison, averaging, and modeling of observed surfaces. Registration is analogous to removing all shape preserving transformations, which include translation, scale, rotation and re-parameterization. This framework is based on a special representation of surfaces termed square-root normal fields and a closely related elastic metric. The main advantages of this method are: (1) the elastic metric provides a natural interpretation of shape deformations that are being quantified, (2) this metric is invariant to re-parameterizations of surfaces, and (3) under the square-root normal field transformation, the complicated elastic metric becomes the standard L2 metric, simplifying parts of the implementation. We present numerous examples of shape comparisons for various types of surfaces in different application areas. We also compute average shapes, covariances and perform principal component analysis to explore the variability in different shape classes. These quantities are used to define generative shape models and for random sampling. Specifically, we showcase the applicability of the proposed framework in shape analysis of anatomical structures in different medical applications including Attention Deficit Hyperactivity Disorder and endometriosis.
Sebastian KurtekWe present a new Riemannian framework for comprehensive statistical shape analysis of 3D objects, represented by their boundaries (parameterized surfaces). By comprehensive framework, we mean tools for registration, comparison, averaging, and modeling of observed surfaces. Registration is analogous to removing all shape preserving transformations, which include translation, scale, rotation and re-parameterization. This framework is based on a special representation of surfaces termed square-root normal fields and a closely related elastic metric. The main advantages of this method are: (1) the elastic metric provides a natural interpretation of shape deformations that are being quantified, (2) this metric is invariant to re-parameterizations of surfaces, and (3) under the square-root normal field transformation, the complicated elastic metric becomes the standard L2 metric, simplifying parts of the implementation. We present numerous examples of shape comparisons for various types of surfaces in different application areas. We also compute average shapes, covariances and perform principal component analysis to explore the variability in different shape classes. These quantities are used to define generative shape models and for random sampling. Specifically, we showcase the applicability of the proposed framework in shape analysis of anatomical structures in different medical applications including Attention Deficit Hyperactivity Disorder and endometriosis. Wojciech ChacholskiI will describe a method of constructing continuous invariants of multidimensional persistence modules using so called noise systems. My aim for this talk is to discuss questions related to stability and computability of such invariants.
Wojciech ChacholskiI will describe a method of constructing continuous invariants of multidimensional persistence modules using so called noise systems. My aim for this talk is to discuss questions related to stability and computability of such invariants.