-

Dane Taylor
Social and biological contagions are influenced by the spatial embeddedness of networks. Historically, many epidemics spread as a wave across part of the Earth's surface; however, in modern contagions long-range edges -- for example, due to airline transportation or communication media -- allow clusters of a contagion to appear in distant locations. Here we study the spread of contagions on networks through a methodology grounded in topological data analysis and nonlinear dimension reduction. We construct "contagion maps" that use multiple contagions on a network to map the nodes as a point cloud. By analyzing the topology, geometry, and dimensionality of manifold structure in such point clouds, we reveal insights to aid in the modeling, forecast, and control of spreading processes. Our approach highlights contagion maps also as a viable tool for inferring low-dimensional structure in networks.
-

Chiara Poletto
Our understanding of communicable disease prevention and control is rooted in the theory of host population transmission dynamics. To this end, the network of host-to-host contacts along which the transmission can occur drives the disease spreading. Network theory provides, thus, tools for assessing the potential for sustained spreading, and understanding the pattern and the velocity of the latter.
Recently different types of sensors and devices allow mining highly resolved host contact data in space and time in different contexts relevant for disease spread. Data availability reveals the temporal dimension of such systems and pushes the development of new approaches for the identification of the temporal aspects of the contact patterns that are critical for disease transmission.
The talk will focus on risk assessment analyses based on the identification of the epidemic threshold, the critical infection transmissibility value separating disease extinction from sustained spreading. I will present a theoretical framework, the infection propagator approach, able to assess this quantity for the case of host-to-host contact networks that are variable in time [1,2]. The approach relays on the knowledge of the sequence of adjacency matrices describing the temporal network and is rooted in a multi-layer representation of the latter that preserves the network causality. It employs a tensor formulation that integrates both spreading and network dynamics and allows for the analytical solution of the linearized Markov chain description of the spreading process, extending in this way the quenched approach to the time-varying case.
I will discuss the mathematical formulation of the approach for various disease compartmental schemes, SIS, SIR, SIRS [1,2] and SEIS models and I will present examples of how this can be applied to epidemiological situations of interest to provide insights on the factors driving the epidemic risk.
[1] Analytical computation of the epidemic threshold on temporal networks E. Valdano, L. Ferreri, C. Poletto, V. Colizza, Physics Review X 5 021005 (2015)
[2] Infection propagator approach to compute epidemic thresholds on temporal networks: impact of immunity and of limited temporal resolution, E. Valdano, C. Poletto, V. Colizza, the European Physical Journal B, 88: 341 (2015)
-

-

-

-

-

-

Blair Sullivan
-

Peter Mucha
A prominent problem in the application of networks to various disciplines is the algorithmic detection of tightly connected groups of nodes known as communities. Recently, there has been increased interest in networks with multiple types of relationships, that change in time, or that network together multiple kinds of networks. We describe different ways that are available for appropriate handling of such "multilayer" features in identifying communities, demonstrating examples with real-world data where such methods have provided new insights.
-
Mikko Kivela
Network science has been very successful in investigations of a wide variety of applications from biology and the social sciences to physics, technology, and more. In many situations, it is already insightful to use a simple (and typically naive) representation as a simple, binary graph in which nodes are entities and unweighted edges encapsulate the interactions between those entities. This allows one to use the powerful methods and concepts for example from graph theory, and numerous advances have been made in this way. However, as network science has matured and (especially) as ever more complicated data has become available, it has become increasingly important to develop tools to analyse more complicated structures. For example, many systems that were typically initially studied as simple graphs are now often represented as time-dependent networks, networks with multiple types of connections, or interdependent networks. This has allowed deeper and more realistic analyses of complex networked systems, but it has simultaneously introduced mathematical constructions, jargon, and methodology that are specific to research in each type of system. Recently, the concept of "multilayer networks" was developed in order to unify the aforementioned disparate language (and disparate notation) and to bring together the different generalised network concepts that included layered graphical structures. In this talk, I will introduce multilayer networks and discuss how to study them.
-
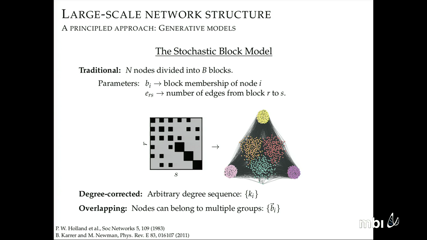
Tiago de Paula Peixoto
To reveal the mechanisms that shape the dynamics on and formation of complex systems, researchers use community-detection methods to describe large-scale patterns in their networks of interactions. Only recently have researchers proposed methods that capture essential memory effects in the dynamics and temporal changes in the formation. However, current memory methods are limited to second-order Markov chain models and current temporal methods are limited to static descriptions in time windows of continuous changes. These limitations raise fundamental questions: how much memory is required and how can time binning be evaded for efficient descriptions based on statistical evidence?
We propose a dynamical description of large-scale structures in sequences and temporal networks that detects the most regularity in the data without any time binning. Our principled approach is based on the statistical inference of generative models, and generalizes the stochastic block model to edge placement probabilities that vary in time and follow an arbitrary-order hidden Markov chain. The method is fully nonparametric and can be used to detect the appropriate Markov order from data alone as well as the number of communities, without overfitting. The method can also predict future network evolution from past observations.
-

Saray Shai
Abstract not submitted
-

Marty Golubitsky
A coupled cell system is a network of interacting dynamical systems. Coupled cell models assume that the output from each node is important and that signals from two or more nodes can be compared so that patterns of synchrony can emerge. The principal question is: How does network architecture (who is talking to whom) affect the kinds of synchronous solutions that are expected in network equations. This talk will discuss a classification of rigid phase-shift synchrony in time-periodic solutions in such systems, as well as some curious synchrony-breaking bifurcations.
-

Tanya Berger-Wolf
From gene interactions and brain activity to cellphone calls and zebras grazing together, large, noisy, and highly dynamic networks of interactions are everywhere. Unfortunately, in this domain, our ability to analyze data lags substantially behind our ability to collect it. Moreover, we may be collecting the wrong data for the questions we want to answer in the first place. From collecting the data and inferring the networks to producing meaningful insight at scale, challenges are there every step of the way and computational approaches have been developed to meet those challenges.
In this talk I will show computational approaches that address some of the questions about dynamic interaction networks: whom should we sample? how often? what is the "right" implicit network? what are the meaningful patterns and trends? and how can we use the network to gain insight into other aspects of the node behavior? The methods leverage the topological graph structure of the networks and the size of the available data to, somewhat counter-intuitively, to produce more accurate results faster. We will demonstrate the scientific implications of the computational analysis on networks of zebras, baboons, and interacting brains cells.
-

Alex Arenas
Most natural and engineered systems include multiple subsystems that are organized as layers of connectivity. It is important to take such features into account to try to improve our understanding of these complex systems. It is thus necessary to generalize "traditional" network theory by developing (and validating) a framework and associated tools to study multilayer systems in a comprehensive fashion and the physical properties that emerge in this new setup. The origins of such efforts date back several decades and arose in multiple disciplines, and now the study of multilayer networks has become one of the most important directions in network science.
Multilayered interconnected networks, and in particular what has been calledmultiplex networks, explicitly incorporate multiple (and independent) channels of connectivity in a system. They provide a natural description for systems in which entities have a different set of neighbors in each layer (which can represent, e.g., a task, an activity, or a category). A fundamental aspect of describing multiplex networks is defining and quantifying the interconnectivity between different categories of connections. This amounts to switching between layers in a multilayer system, and the associated interlayer connections in a network are responsible for the emergence of new phenomena in multiplex networks. Interlayer connections can generate new structural and dynamical correlations between components of a system, so it is important to develop a framework that takes them into account. The study of these systems is of utmost importance to understand more realistic structured interactions.
-

Arkady Pikovsky
-

David Spivak
Rather than considering each node in a network as an atomic entity of some sort, it is often useful to regard the node as potentially consisting of a networked system in its own right. In this talk I will propose the mathematical theory of operads as a foundation for modeling the sort of nested structures that arise when considering the relationship between wholes and parts in a system of systems. The main case of interest will be open dynamical systems (e.g., ODEs or boolean networks), which can be interconnected together to form larger-scale dynamical systems. I will explain how operads can help us more easily calculate invariants of such systems, such as their steady state matrices (which generalize bifurcation diagrams). In particular, I will discuss how the steady state matrix of an interconnected system can be calculated using standard matrix operations (multiplication, tensor product, and trace) applied to the steady state matrices of its component systems.
-

Cristina Masoller
Systems composed by interacting dynamical elements are ubiquitous in nature. In many situations, such systems are modeled as networks of simple oscillators, where the nodes represent the individual units and the links represent the interactions among them. These interactions are often unknown, and a popular method for inferring the underlying connectivity of a system (i.e., the set of links among pairs of nodes) is based on a statistical similarity analysis of the time-series collected from the dynamics of the nodes. In this presentation I will discuss our recent work on inferring network connectivity from observed data. First, I will consider synthetic data and experimental data generated from simple dynamical units (Logistic maps, Kuramoto phase oscillators and Rössler electronic oscillators), which are coupled with known network topology [1, 2]. I will show that, under adequate conditions, the coupling links can be perfectly inferred, i.e., no mistakes are made regarding the presence or absence of links. Then, I will present ongoing work in assessing climate interactions from the analysis of observed climatological data (surface air temperature), recorded at a regular grid of geographical locations covering the Earth surface
[1] N. Rubido, A. C. Marti, E. Bianco-Martinez, C. Grebogi, M. S. Baptista and C. Masoller, "Exact detection of direct links in networks of coupled maps", New Journal of Physics 16 093010 (2014).
[2] G. Tirabassi, R. Sevilla-Escoboza, J. M. Buldú and C. Masoller, “Inferring the connectivity of coupled oscillators from time-series statistical similarity analysis�, Sci. Rep. 5 10829 (2015).
[3] J. I. Deza, M. Barreiro, and C. Masoller, “Assessing the direction of climate interactions by means of complex networks and information theoretic tools�, Chaos 25, 033105 (2015).
-

Georgi Medvedev
We will discuss dynamics in two representative models of dynamical systems on graphs: coupled chaotic maps and coupled excitable systems driven by white noise. For both models, we analyze synchronization and explain what structural features of the network favor synchronization. For the second model, we also describe other dynamical regimes such as spontaneous oscillations and formation of clusters. The role of network topology in shaping each of these patterns will be explained.
-

Ginestra Bianconi
Simplicial complexes are generalized network structures able to encode interactions occurring between more than two nodes. Simplicial complex describe a large variety of complex interacting systems ranging from brain networks, to social and collaboration networks. Additionally simplicial complexes have a geometrical interpretation and for this reasons they have been widely used in quantum gravity. Simplicial complexes are the ideal structures to characterize emergent network geometry in which geometrical properties of the networks emerge spontaneously from their dynamics. Here we propose a general model for growing simplicial complexes called network geometry with flavor (NGF) and we characterize the configuration model of simplicial complexes. These models deepens our understanding of complex networks and reveals the important effect that the dimensionality of growing simplicial complexes have on their structure. The NGF can generate discrete geometries of different nature, ranging from chains and higher dimensional manifolds to scale-free networks with small-world properties, scale-free degree distribution and non-trivial community structure. We find that, for NGF with dimension greater than one, scale-free topologies emerge also without including an explicit preferential attachment because and efficient preferential attachment mechanism naturally emerges from the dynamical rules. Interestingly the NGF with fitness of the nodes reveals relevant relations with quantum statistics. The configuration model of simplicial complexes characterizes instead static simplicial complexes. Here the ensemble will be discussed highlighting the differences with the configuration model of networks, showing that the effect of the increased dimesionality of simplicial complexes is reflected in the different structural cutoff and in the specific nature of the degree correlations.
-

Jennifer Neville
There has been a growing interest in analyzing the network structure of complex systems to understand key patterns/dependencies in the underlying system. This has fueled a large body of research on both models of network structure and algorithms to automatically discover patterns for use in predictive models. However, robust statistical models, which can accurately represent distributions over graph populations, are critical to assess the significance of discovered patterns or to distinguish between alternative models. Moreover, efficient sampling and inference algorithms are crucial for tractable analysis in large-scale domains evolving over time. However, unlike metric spaces, the space of graphs exhibits a combinatorial structure that poses significant theoretical and practical challenges to accurate estimation and efficient sampling/inference. In this talk, I will discuss our recent work on modeling distributions of networks and outline how the methods can be used for hypothesis testing, anomaly detection, and anonymization.
 Dane TaylorSocial and biological contagions are influenced by the spatial embeddedness of networks. Historically, many epidemics spread as a wave across part of the Earth's surface; however, in modern contagions long-range edges -- for example, due to airline transportation or communication media -- allow clusters of a contagion to appear in distant locations. Here we study the spread of contagions on networks through a methodology grounded in topological data analysis and nonlinear dimension reduction. We construct "contagion maps" that use multiple contagions on a network to map the nodes as a point cloud. By analyzing the topology, geometry, and dimensionality of manifold structure in such point clouds, we reveal insights to aid in the modeling, forecast, and control of spreading processes. Our approach highlights contagion maps also as a viable tool for inferring low-dimensional structure in networks.
Dane TaylorSocial and biological contagions are influenced by the spatial embeddedness of networks. Historically, many epidemics spread as a wave across part of the Earth's surface; however, in modern contagions long-range edges -- for example, due to airline transportation or communication media -- allow clusters of a contagion to appear in distant locations. Here we study the spread of contagions on networks through a methodology grounded in topological data analysis and nonlinear dimension reduction. We construct "contagion maps" that use multiple contagions on a network to map the nodes as a point cloud. By analyzing the topology, geometry, and dimensionality of manifold structure in such point clouds, we reveal insights to aid in the modeling, forecast, and control of spreading processes. Our approach highlights contagion maps also as a viable tool for inferring low-dimensional structure in networks. Chiara PolettoOur understanding of communicable disease prevention and control is rooted in the theory of host population transmission dynamics. To this end, the network of host-to-host contacts along which the transmission can occur drives the disease spreading. Network theory provides, thus, tools for assessing the potential for sustained spreading, and understanding the pattern and the velocity of the latter.
Chiara PolettoOur understanding of communicable disease prevention and control is rooted in the theory of host population transmission dynamics. To this end, the network of host-to-host contacts along which the transmission can occur drives the disease spreading. Network theory provides, thus, tools for assessing the potential for sustained spreading, and understanding the pattern and the velocity of the latter.




 Blair Sullivan
Blair Sullivan Peter MuchaA prominent problem in the application of networks to various disciplines is the algorithmic detection of tightly connected groups of nodes known as communities. Recently, there has been increased interest in networks with multiple types of relationships, that change in time, or that network together multiple kinds of networks. We describe different ways that are available for appropriate handling of such "multilayer" features in identifying communities, demonstrating examples with real-world data where such methods have provided new insights.
Peter MuchaA prominent problem in the application of networks to various disciplines is the algorithmic detection of tightly connected groups of nodes known as communities. Recently, there has been increased interest in networks with multiple types of relationships, that change in time, or that network together multiple kinds of networks. We describe different ways that are available for appropriate handling of such "multilayer" features in identifying communities, demonstrating examples with real-world data where such methods have provided new insights. Mikko KivelaNetwork science has been very successful in investigations of a wide variety of applications from biology and the social sciences to physics, technology, and more. In many situations, it is already insightful to use a simple (and typically naive) representation as a simple, binary graph in which nodes are entities and unweighted edges encapsulate the interactions between those entities. This allows one to use the powerful methods and concepts for example from graph theory, and numerous advances have been made in this way. However, as network science has matured and (especially) as ever more complicated data has become available, it has become increasingly important to develop tools to analyse more complicated structures. For example, many systems that were typically initially studied as simple graphs are now often represented as time-dependent networks, networks with multiple types of connections, or interdependent networks. This has allowed deeper and more realistic analyses of complex networked systems, but it has simultaneously introduced mathematical constructions, jargon, and methodology that are specific to research in each type of system. Recently, the concept of "multilayer networks" was developed in order to unify the aforementioned disparate language (and disparate notation) and to bring together the different generalised network concepts that included layered graphical structures. In this talk, I will introduce multilayer networks and discuss how to study them.
Mikko KivelaNetwork science has been very successful in investigations of a wide variety of applications from biology and the social sciences to physics, technology, and more. In many situations, it is already insightful to use a simple (and typically naive) representation as a simple, binary graph in which nodes are entities and unweighted edges encapsulate the interactions between those entities. This allows one to use the powerful methods and concepts for example from graph theory, and numerous advances have been made in this way. However, as network science has matured and (especially) as ever more complicated data has become available, it has become increasingly important to develop tools to analyse more complicated structures. For example, many systems that were typically initially studied as simple graphs are now often represented as time-dependent networks, networks with multiple types of connections, or interdependent networks. This has allowed deeper and more realistic analyses of complex networked systems, but it has simultaneously introduced mathematical constructions, jargon, and methodology that are specific to research in each type of system. Recently, the concept of "multilayer networks" was developed in order to unify the aforementioned disparate language (and disparate notation) and to bring together the different generalised network concepts that included layered graphical structures. In this talk, I will introduce multilayer networks and discuss how to study them. Tiago de Paula PeixotoTo reveal the mechanisms that shape the dynamics on and formation of complex systems, researchers use community-detection methods to describe large-scale patterns in their networks of interactions. Only recently have researchers proposed methods that capture essential memory effects in the dynamics and temporal changes in the formation. However, current memory methods are limited to second-order Markov chain models and current temporal methods are limited to static descriptions in time windows of continuous changes. These limitations raise fundamental questions: how much memory is required and how can time binning be evaded for efficient descriptions based on statistical evidence?
Tiago de Paula PeixotoTo reveal the mechanisms that shape the dynamics on and formation of complex systems, researchers use community-detection methods to describe large-scale patterns in their networks of interactions. Only recently have researchers proposed methods that capture essential memory effects in the dynamics and temporal changes in the formation. However, current memory methods are limited to second-order Markov chain models and current temporal methods are limited to static descriptions in time windows of continuous changes. These limitations raise fundamental questions: how much memory is required and how can time binning be evaded for efficient descriptions based on statistical evidence? Saray ShaiAbstract not submitted
Saray ShaiAbstract not submitted Marty GolubitskyA coupled cell system is a network of interacting dynamical systems. Coupled cell models assume that the output from each node is important and that signals from two or more nodes can be compared so that patterns of synchrony can emerge. The principal question is: How does network architecture (who is talking to whom) affect the kinds of synchronous solutions that are expected in network equations. This talk will discuss a classification of rigid phase-shift synchrony in time-periodic solutions in such systems, as well as some curious synchrony-breaking bifurcations.
Marty GolubitskyA coupled cell system is a network of interacting dynamical systems. Coupled cell models assume that the output from each node is important and that signals from two or more nodes can be compared so that patterns of synchrony can emerge. The principal question is: How does network architecture (who is talking to whom) affect the kinds of synchronous solutions that are expected in network equations. This talk will discuss a classification of rigid phase-shift synchrony in time-periodic solutions in such systems, as well as some curious synchrony-breaking bifurcations. Tanya Berger-WolfFrom gene interactions and brain activity to cellphone calls and zebras grazing together, large, noisy, and highly dynamic networks of interactions are everywhere. Unfortunately, in this domain, our ability to analyze data lags substantially behind our ability to collect it. Moreover, we may be collecting the wrong data for the questions we want to answer in the first place. From collecting the data and inferring the networks to producing meaningful insight at scale, challenges are there every step of the way and computational approaches have been developed to meet those challenges.
Tanya Berger-WolfFrom gene interactions and brain activity to cellphone calls and zebras grazing together, large, noisy, and highly dynamic networks of interactions are everywhere. Unfortunately, in this domain, our ability to analyze data lags substantially behind our ability to collect it. Moreover, we may be collecting the wrong data for the questions we want to answer in the first place. From collecting the data and inferring the networks to producing meaningful insight at scale, challenges are there every step of the way and computational approaches have been developed to meet those challenges. Alex ArenasMost natural and engineered systems include multiple subsystems that are organized as layers of connectivity. It is important to take such features into account to try to improve our understanding of these complex systems. It is thus necessary to generalize "traditional" network theory by developing (and validating) a framework and associated tools to study multilayer systems in a comprehensive fashion and the physical properties that emerge in this new setup. The origins of such efforts date back several decades and arose in multiple disciplines, and now the study of multilayer networks has become one of the most important directions in network science.
Alex ArenasMost natural and engineered systems include multiple subsystems that are organized as layers of connectivity. It is important to take such features into account to try to improve our understanding of these complex systems. It is thus necessary to generalize "traditional" network theory by developing (and validating) a framework and associated tools to study multilayer systems in a comprehensive fashion and the physical properties that emerge in this new setup. The origins of such efforts date back several decades and arose in multiple disciplines, and now the study of multilayer networks has become one of the most important directions in network science. Arkady Pikovsky
Arkady Pikovsky David SpivakRather than considering each node in a network as an atomic entity of some sort, it is often useful to regard the node as potentially consisting of a networked system in its own right. In this talk I will propose the mathematical theory of operads as a foundation for modeling the sort of nested structures that arise when considering the relationship between wholes and parts in a system of systems. The main case of interest will be open dynamical systems (e.g., ODEs or boolean networks), which can be interconnected together to form larger-scale dynamical systems. I will explain how operads can help us more easily calculate invariants of such systems, such as their steady state matrices (which generalize bifurcation diagrams). In particular, I will discuss how the steady state matrix of an interconnected system can be calculated using standard matrix operations (multiplication, tensor product, and trace) applied to the steady state matrices of its component systems.
David SpivakRather than considering each node in a network as an atomic entity of some sort, it is often useful to regard the node as potentially consisting of a networked system in its own right. In this talk I will propose the mathematical theory of operads as a foundation for modeling the sort of nested structures that arise when considering the relationship between wholes and parts in a system of systems. The main case of interest will be open dynamical systems (e.g., ODEs or boolean networks), which can be interconnected together to form larger-scale dynamical systems. I will explain how operads can help us more easily calculate invariants of such systems, such as their steady state matrices (which generalize bifurcation diagrams). In particular, I will discuss how the steady state matrix of an interconnected system can be calculated using standard matrix operations (multiplication, tensor product, and trace) applied to the steady state matrices of its component systems. Cristina MasollerSystems composed by interacting dynamical elements are ubiquitous in nature. In many situations, such systems are modeled as networks of simple oscillators, where the nodes represent the individual units and the links represent the interactions among them. These interactions are often unknown, and a popular method for inferring the underlying connectivity of a system (i.e., the set of links among pairs of nodes) is based on a statistical similarity analysis of the time-series collected from the dynamics of the nodes. In this presentation I will discuss our recent work on inferring network connectivity from observed data. First, I will consider synthetic data and experimental data generated from simple dynamical units (Logistic maps, Kuramoto phase oscillators and Rössler electronic oscillators), which are coupled with known network topology [1, 2]. I will show that, under adequate conditions, the coupling links can be perfectly inferred, i.e., no mistakes are made regarding the presence or absence of links. Then, I will present ongoing work in assessing climate interactions from the analysis of observed climatological data (surface air temperature), recorded at a regular grid of geographical locations covering the Earth surface
Cristina MasollerSystems composed by interacting dynamical elements are ubiquitous in nature. In many situations, such systems are modeled as networks of simple oscillators, where the nodes represent the individual units and the links represent the interactions among them. These interactions are often unknown, and a popular method for inferring the underlying connectivity of a system (i.e., the set of links among pairs of nodes) is based on a statistical similarity analysis of the time-series collected from the dynamics of the nodes. In this presentation I will discuss our recent work on inferring network connectivity from observed data. First, I will consider synthetic data and experimental data generated from simple dynamical units (Logistic maps, Kuramoto phase oscillators and Rössler electronic oscillators), which are coupled with known network topology [1, 2]. I will show that, under adequate conditions, the coupling links can be perfectly inferred, i.e., no mistakes are made regarding the presence or absence of links. Then, I will present ongoing work in assessing climate interactions from the analysis of observed climatological data (surface air temperature), recorded at a regular grid of geographical locations covering the Earth surface Georgi MedvedevWe will discuss dynamics in two representative models of dynamical systems on graphs: coupled chaotic maps and coupled excitable systems driven by white noise. For both models, we analyze synchronization and explain what structural features of the network favor synchronization. For the second model, we also describe other dynamical regimes such as spontaneous oscillations and formation of clusters. The role of network topology in shaping each of these patterns will be explained.
Georgi MedvedevWe will discuss dynamics in two representative models of dynamical systems on graphs: coupled chaotic maps and coupled excitable systems driven by white noise. For both models, we analyze synchronization and explain what structural features of the network favor synchronization. For the second model, we also describe other dynamical regimes such as spontaneous oscillations and formation of clusters. The role of network topology in shaping each of these patterns will be explained. Ginestra BianconiSimplicial complexes are generalized network structures able to encode interactions occurring between more than two nodes. Simplicial complex describe a large variety of complex interacting systems ranging from brain networks, to social and collaboration networks. Additionally simplicial complexes have a geometrical interpretation and for this reasons they have been widely used in quantum gravity. Simplicial complexes are the ideal structures to characterize emergent network geometry in which geometrical properties of the networks emerge spontaneously from their dynamics. Here we propose a general model for growing simplicial complexes called network geometry with flavor (NGF) and we characterize the configuration model of simplicial complexes. These models deepens our understanding of complex networks and reveals the important effect that the dimensionality of growing simplicial complexes have on their structure. The NGF can generate discrete geometries of different nature, ranging from chains and higher dimensional manifolds to scale-free networks with small-world properties, scale-free degree distribution and non-trivial community structure. We find that, for NGF with dimension greater than one, scale-free topologies emerge also without including an explicit preferential attachment because and efficient preferential attachment mechanism naturally emerges from the dynamical rules. Interestingly the NGF with fitness of the nodes reveals relevant relations with quantum statistics. The configuration model of simplicial complexes characterizes instead static simplicial complexes. Here the ensemble will be discussed highlighting the differences with the configuration model of networks, showing that the effect of the increased dimesionality of simplicial complexes is reflected in the different structural cutoff and in the specific nature of the degree correlations.
Ginestra BianconiSimplicial complexes are generalized network structures able to encode interactions occurring between more than two nodes. Simplicial complex describe a large variety of complex interacting systems ranging from brain networks, to social and collaboration networks. Additionally simplicial complexes have a geometrical interpretation and for this reasons they have been widely used in quantum gravity. Simplicial complexes are the ideal structures to characterize emergent network geometry in which geometrical properties of the networks emerge spontaneously from their dynamics. Here we propose a general model for growing simplicial complexes called network geometry with flavor (NGF) and we characterize the configuration model of simplicial complexes. These models deepens our understanding of complex networks and reveals the important effect that the dimensionality of growing simplicial complexes have on their structure. The NGF can generate discrete geometries of different nature, ranging from chains and higher dimensional manifolds to scale-free networks with small-world properties, scale-free degree distribution and non-trivial community structure. We find that, for NGF with dimension greater than one, scale-free topologies emerge also without including an explicit preferential attachment because and efficient preferential attachment mechanism naturally emerges from the dynamical rules. Interestingly the NGF with fitness of the nodes reveals relevant relations with quantum statistics. The configuration model of simplicial complexes characterizes instead static simplicial complexes. Here the ensemble will be discussed highlighting the differences with the configuration model of networks, showing that the effect of the increased dimesionality of simplicial complexes is reflected in the different structural cutoff and in the specific nature of the degree correlations. Jennifer NevilleThere has been a growing interest in analyzing the network structure of complex systems to understand key patterns/dependencies in the underlying system. This has fueled a large body of research on both models of network structure and algorithms to automatically discover patterns for use in predictive models. However, robust statistical models, which can accurately represent distributions over graph populations, are critical to assess the significance of discovered patterns or to distinguish between alternative models. Moreover, efficient sampling and inference algorithms are crucial for tractable analysis in large-scale domains evolving over time. However, unlike metric spaces, the space of graphs exhibits a combinatorial structure that poses significant theoretical and practical challenges to accurate estimation and efficient sampling/inference. In this talk, I will discuss our recent work on modeling distributions of networks and outline how the methods can be used for hypothesis testing, anomaly detection, and anonymization.
Jennifer NevilleThere has been a growing interest in analyzing the network structure of complex systems to understand key patterns/dependencies in the underlying system. This has fueled a large body of research on both models of network structure and algorithms to automatically discover patterns for use in predictive models. However, robust statistical models, which can accurately represent distributions over graph populations, are critical to assess the significance of discovered patterns or to distinguish between alternative models. Moreover, efficient sampling and inference algorithms are crucial for tractable analysis in large-scale domains evolving over time. However, unlike metric spaces, the space of graphs exhibits a combinatorial structure that poses significant theoretical and practical challenges to accurate estimation and efficient sampling/inference. In this talk, I will discuss our recent work on modeling distributions of networks and outline how the methods can be used for hypothesis testing, anomaly detection, and anonymization.